Isolation and Preliminary Characterization of a Novel scFv against SARS-CoV-2: an Experimental and Computational Analysis
-
Jahandar-Lashaki, Samaneh
-
Department of Medical Biotechnology, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
-
 Farajnia, Safar
Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran , Tel: +98 9143018589; Fax: +98 41 33363432; E-mail: farajnias@tbzmed.ac.ir
Farajnia, Safar
Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran , Tel: +98 9143018589; Fax: +98 41 33363432; E-mail: farajnias@tbzmed.ac.ir
-
Biotechnology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
-
Alizadeh , Effat
-
Department of Medical Biotechnology, Faculty of Advanced Medical Sciences, Tabriz University of Medical Sciences, Tabriz, Iran
-
Seirafi, Farzin
-
Biotechnology Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
-
Tanoumand, Asghar
-
Department of Microbiology, Maragheh University of Medical Sciences, Maragheh, Maragheh, Iran
Abstract: Background: Since the initial outbreak, the SARS-CoV-2 virus has continued to circulate and mutate, resulting in the emergence of new viral sublineages. Due to the lack of effective protection and therapeutic measures against these new variants, the virus is able to further evolve and diversify. This study aimed to screen a phage antibody library to identify monoclonal antibodies in single-chain variable fragment (scFv) format that target the Receptor Binding Domain (RBD) of different SARS-CoV-2 strains. The newly discovered scFv has the potential for use as a diagnostic or therapeutic option against SARS-CoV-2.
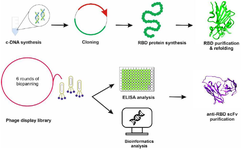
Methods: The RBD protein was produced, purified, and used as an antigen during biopanning. Six rounds of panning enriched RBD-specific phages and the binding affinity of binders were monitored by polyclonal phage ELISA. Subsequently, monoclonal phage ELISA was employed to identify specific binders. After sequence confirmation, the reactivity of the isolated anti-RBD scFv was evaluated. Additionally, bioinformatics tools determined the interaction between selected scFv and SARS-CoV-2 strains.
Results: The ELISA analysis demonstrated that the expressed RBD retains its structural integrity and effectively interacts with antibodies present in the sera of COVID-19 patients. Through screening a phage display library, a strong-binding scFv for RBD was discovered, which can effectively neutralize SARS-CoV-2 and its novel variants.
Conclusion: The findings of this study have led to the discovery of a novel scFv that effectively neutralizes SARS-CoV-2 strains, offering immense potential for research and therapy purposes.
Introduction :
The global health landscape was profoundly impacted by the unprecedented COVID-19 pandemic, caused by the highly contagious SARS-CoV-2 virus 1. Currently, the SARS-CoV-2 Spike protein is undergoing continuous mutations, leading to the emergence of novel variants known as Variants Of Interest (VOIs) and Variants Under Monitoring (VUMs) such as XBB and BA.2 lineages 2. These variants are responsible for breakthrough infections in vaccinated individuals and can reduce the effectiveness of therapeutic interventions. Additionally, it is anticipated that SARS-CoV-2 will remain in circulation for a prolonged period, much like other viruses that have caused pandemics in the past. Therefore, the development of alternative therapeutics for treating patients with severe clinical symptoms remains a priority 3,4.
SARS-CoV-2, a member of the Coronaviridae family, is an enveloped virus classified under the betacoronavirus genus. The positive-sense single-stranded RNA [(+) ssRNA] genome of the virus encodes four structural proteins (Spike, Membrane, Nucleocapsid, and Envelope protein) 5. Among the structural proteins, spike glycoprotein, which is found on the viral envelope, plays a dominant role in viral entry. The transmembrane Spike (S) protein is composed of two subunits, S1 and S2, with distinct functions. S1 is responsible for receptor binding, while S2 facilitates the fusion of viral and cellular membranes 6. The Receptor-Binding Domain (RBD) located within the S1 subunit mediates binding to the Angiotensin-Converting Enzyme 2 (ACE2) receptor. Following the ACE2-RBD interaction, conformational changes in the S2 subunit leads to viral entry 7.
Some people, including those receiving chemotherapy, those with hematologic malignancies and immunocompromised individuals, may not benefit from COVID-19 vaccines 8,9. Additionally, there are limited options for COVID-19 treatment. Thus, a novel therapeutic approach is needed to manage the disease and improve patient survival rates. Recently, several therapeutic strategies have been developed, including inflammatory modulators, antiviral drugs, stem cell therapies, convalescent plasma treatments, and, lastly, antibody therapies 10. Among these strategies, antibodies are the most promising approach for disease prevention and treatment, given their success in previous research 11-13. Monoclonal antibodies (mAbs) such as Etesevimab and Bamlanivimab have been authorized for emergency use in the treatment of COVID-19. These antibodies are designed to block the interaction between the viral spike protein and the ACE2 receptor, effectively neutralizing the virus 14. However, the rapid emergence of SARS-CoV-2 variants with mutations in the spike protein has raised concerns about the long-term efficacy of these mAbs, as some variants have shown reduced susceptibility to neutralization 15.
The discovery and development of antibodies can be achieved using a variety of approaches. One method is phage display libraries, which have been widely used to screen for human antibodies 16. Phage display technology involves fusing millions of peptide sequences with phage proteins and displaying them on the phage surface. Phenotype-genotype linkages, as well as specific screening for target antigens based on binding affinity, are essential aspects of this approach 17. Since phage display screening is performed in vitro, therapeutic antibodies can be isolated in various settings 18. In phage libraries, the predominant antibody formats utilized are antigen-binding fragments (Fabs) and single-chain variable fragments (scFvs). Fabs consist of both variable domains (VL and VH) and constant domains (CL and CH1), whereas scFvs consist solely of the VH and VL domains that are linked through a flexible linker 19.
The development of antibodies against SARS-CoV-2 has been extensively studied. Most neutralizing antibodies derived from COVID-19 patients target the RBD of the S protein, demonstrating its high immunogenicity 20. In this study, the Human Single Fold scFv Library (Tomlinson I) against the RBD of the SARS-CoV-2 S protein was screened in order to isolate a human anti-RBD scFv as a candidate for further development in the ongoing search for effective therapeutic antibodies against SARS-CoV-2.
Materials and Methods :
Phage library and bacterial strains: The Medical Research Council (MRC) in Cambridge provided the semisynthetic human scFv phage libraries I & J (Tomlinson I& J), KM13 helper phage, and Escherichia coli (E. coli) (TG1) strain. Novagen supplied E. coli BL21(DE3).
Complementary DNA (cDNA) synthesis: RNA samples of the SARS-COV-2 virus were extracted from samples of hospitalized patients and sent to the laboratory. The extracted RNA was utilized for cDNA synthesis by SinaClone cDNA synthesis kit (Cat. No.: RT5201) as described by the manufacturer protocol. Finally, the quality of the cDNA was assessed using a Nanodrop spectrophotometer (260/280 nm).
Polymerase chain reaction (PCR) and DNA sequencing: Polymerase Chain Reaction (PCR) was carried out to amplify the RBD sequence of the SARS-CoV-2 S protein. The reaction was performed on the cDNA and the primer sets contained Nde I and Xho I restriction sites (forward: ATACATATGAGAGGTGATGAAG TC; reverse: GTGCTCGAGTTAAACAGTTGCTG GT). The purification of the RBD PCR product was carried out by using the GEL/PCR Purification Kit (FAVORGEN, Cat. No.: FAGCK001) subsequent to its separation on a 1% agarose gel. To analyze the results of the PCR amplification and DNA extraction, the agarose gel was visualized using exposure to UV radiation. Also, DNA sequencing was accomplished to confirm the final RBD sequence.
Expression, purification, and refolding of the receptor-binding domain of SARS-CoV-2 spike glycoprotein: The E. coli (DH5α) colonies harboring pET28a (+) vector were cultured in LB medium and used for plasmid extraction by Plasmid Extraction Kit (FAVORGEN, Cat. No.: FAPDE 001-1). The pET28a (+) plasmid and the sequence coding for RBD were treated with NdeI and XhoI restriction enzymes at 37℃ for 1 hr, run on 1% agarose gel, and the target bands were purified by GEL/PCR Purification and Plasmid Extraction Kit. Then, the RBD fragment was ligated into pET28a (+) using T4 DNA ligase (Thermofisher, Cat. No. EL0011) based on the manufacturer protocol. The recombinant construct was introduced into the competent E. coli (BL21) and cultured in LB medium containing kanamycin. Subsequently, the culture was induced with a concentration of 0.5 mM IPTG at OD600=0.5 for 16 hr at 30℃ to produce a high yield of recombinant RBD. Finally, the expression was analyzed using 15% SDS-page.
For purification, the culture was centrifuged at 9000 rpm for 30 min. The pellet was sonicated on an ice bath using Tris-Na2HPO4 buffer (pH=8.0). Finally, the pellet containing RBD was harvested by centrifugation at 9000 rpm for 30 min. The recombinant RBD was purified from the insoluble fraction by the Ni-NTA column according to QIAGEN protocol. To facilitate the refolding of the RBD expressed in E. coli, 0.9 mM GSSG and 1.8 mM GSH were used in the refolding buffer for dialysis along with 10 mM DTT, 400 mM L-arginine, and 8 M urea (pH=10). The DTT concentration was gradually reduced while maintaining the arginine concentration. Urea was also removed step by step. Finally, the quality of purified RBD was assessed using SDS-PAGE.
RBD ELISA assay: The integrity of RBD protein was assessed by indirect ELISA. The positive and negative sera of COVID-19 patients were selected from PCR-confirmed samples and IgG-IgM antibody levels. The positive and negative samples were combined separately to prepare positive and negative pools. The 96-well plate was coated with 5 µg/ml RBD and blocked with PBS-T contained 3% BSA. Then positive and negative serum pools (with 1:25, 1:50, 1:100, and 1:200 dilutions) were poured into the wells and incubated at RT for 1 hr. After washing, HRP-conjugated anti-human IgG was added and binding antibodies were visualized by TMB substrate solution.
Phage display library amplification and panning: For library amplification, the Tomlinson I library was cultured in pre-warmed 2xTY media with 100 µg/ml ampicillin and 1% glucose. The culturing process continued until the OD 600 reached 0.4. Following a 30-min period of incubation at a temperature of 37°C, the culture was inoculated in 2×TY medium, which was enriched with 0.1% glucose, 50 µg/ml kanamycin, and 100 µg/ml ampicillin. The culture was then allowed to incubate overnight at 30°C. The next day, PEG/NaCl was added to the centrifuged culture and finally, phages were resuspended in Phosphate Buffered Saline (PBS).
The biopanning procedure and following assays was performed according to the MRC protocol and based on our previous studies 21,22 for six rounds against the RBD protein. Briefly, the process involved coating a Maxisorb 96-well plate with RBD protein (Table 1) at 4℃ overnight, followed by blocking with 2% Marvel skimmed milk powder in PBS (MPBS) at Room Temperature (RT) for 2 hr. After three washes with PBS, a solution containing 1012 to 1013 pfu of phages, diluted in 2% MPBS was added to the plate. The plate was then incubated at RT for a duration of 2 hr. To eliminate non-specific phages, the plate was washed with PBS-0.1% Tween: 10 times during round 1, 20 times for rounds 2-3, and 30 times for subsequent rounds. The bound phages were eluted by adding 100 µl of trypsin-PBS (10 µl of 10 mg/ml trypsin in 90 µl PBS) to each well. A total of 250 µl of eluted phages was added to 1.75 ml of E. coli TG1 (OD 600 of 0.4). After a 30-min incubation in a water bath at 37°C, 100 µl of the TG1 culture was resuspended in 2×TY. The resuspended culture was poured onto a TYE plate containing 1% glucose and 100 µg/ml ampicillin, and the plate was left to grow overnight at 37°C. For subsequent rounds of phage selection, 7 ml of 2×TY medium was added to the overnight culture, and the bacteria were scraped using a glass spreader. Finally, 50 µl of the scraped bacteria was used to purify the phage obtained from each round using PEG/NaCl, as mentioned above.
Polyclonal phage ELISA: Following each cycle of panning, the phage populations were screened for binding to the RBD using a polyclonal phage ELISA to determine the specificity of the acquired binders and to confirm the completion of the biopanning process. For this purpose, 96-well ELISA plates were coated with 10 µg/ml RBD protein and blocked by 3% BSA. After blocking, 10 µl PEG-precipitated phages of each round were added into wells containing 100 µl of 3% Bovine Serum Albumin (BSA). After washing three times with 0.1% Tween 20-PBS, HRP-conjugated anti-M13 antibody diluted in 3% BSA at 1:5000 was added and phages-RBD interactions were detected using TMB as the substrate solution. Absorbance at 450 nm was read as binding signals.
PCR screening of the selected colonies: Based on the polyclonal phage ELISA results over 64 colonies from round 5 were selected and the existence of full-length scFvs was verified by conducting PCR screening on each individual clone using primer sets for VH and Vκ (Table 2).
Monoclonal phage ELISA: The identification of monoclonal phage antibodies was achieved through a screening process utilizing ELISA on selected colonies from round 5. These colonies were subsequently transferred into 96-well plates containing 2×TY medium, enriched with 100 µg/ml ampicillin and 1% glucose. After 2 hr of incubation, 25 µl of 2×TY medium containing 109 helper phage, 1% glucose, and 100 µg/ml ampicillin was added into the wells, incubated for 60 min, and centrifuged. Then, the cells were resuspended in 200 µl of 2×TY, which was enriched with 50 µg/ml kanamycin and 100 µg/ml ampicillin. Following overnight incubation at 30°C, the culture was subjected to centrifugation, and a 50 µl sample of the supernatant was used in phage ELISA, as previously explained.
Generation of antibody fragments: Final selected colonies were induced in TG-1 to express soluble scFv fragment. Individual colonies were cultured in 2×TY containing 1% glucose and 100 µg/ml ampicillin in 96-well plates at 37℃. A small volume (2 µl) of the cultured colonies was then transferred into a second plate containing 2×TY medium enriched with 0.1% glucose and 100 µg/ml ampicillin. After incubating for 3 hr at 37℃, 2×TY medium containing 0.5 mM IPTG and 100 µg/ml ampicillin was poured onto the plate. The plate was then incubated overnight at 30°C. The culture supernatant obtained from this incubation was used for phage ELISA, as described previously, and the binding detection was performed using HRP (horseradish peroxidase)-Protein L.
Antibodies sequence analysis: Selected colonies with high binding affinity and specificity were sequenced using Vκ and VH primers (Table 2).
Selected scFv subcloning, expression and purification: To increase the expression yield, the plasmid was extracted from a positive clone, which had higher reactivity in ELISA. Then, after dilution at a ratio of 1 to 10, PCR reaction was performed. After purification as previously described, the PCR product was used for cloning. Specific primers for this reaction were designed with restriction sites for NdeI and BamHI enzymes. Subsequently, the PCR product and pET28a vector were individually digested in separate microtubes using NdeI and BamHI enzymes at 37℃ for 4 hr. To construct the recombinant vector, the PCR product obtained from the double digestion was ligated into the digested PET28a using T4 DNA ligase (Thermofisher co.). The recombinant construct was transferred into E. coli (DH5a) and cultured in LB medium containing kanamycin. Finally, positive clones containing the recombinant vector were confirmed by PCR reaction.
To assess expression of the selected scFv of E. coli BL21 (DE3), pLysS cells were prepared by calcium chloride method and the recombinant vector was transferred to them by heat shock method and cultured in the LB medium containing kanamycin 23. The positive clones containing recombinant scFv were induced with 1 mM IPTG at 30℃ overnight and the expression was checked using SDS-PAGE.
After optimizing the conditions for antibody expression and determining that the antibody was insoluble, the sediment obtained from sonication was resolved in the lysing buffer (Na2HPO4 100 mM, Tris 10 mM) and 8 M urea. The final volume of the solution was 8 ml. Then, the protein purification was performed using Ni-NTA column according to QIAGEN protocol. Finally, the dissolved scFv molecules were refolded through dialysis to eliminate urea and imidazole, employing a solution containing 10 mM DTT and 200 mM arginine at pH=10.5. The quality of the purification was assessed using SDS-PAGE.
Investigation of anti-RBD activity using Indirect ELISA: For this purpose, ELISA was used to assess the initial binding of the recombinant scFv antibody to the SARS-CoV-2 RBD. The ELISA wells were coated with RBD at a concentration of 40 μg/ml. After washing and blocking, the scFv antibody was added to the wells at different dilutions and left to incubate at RT for one hr. Then, the wells were washed using PBS-T buffer, which was repeated five times. Then, L-HRP was added at a dilution ratio of 1:2000, and the wells were left to incubate at RT for an hr. Finally, the reaction was visualized by adding TMB substrate, and the absorbance was subsequently quantified at a wavelength of 450 nm utilizing an ELISA reader.
scFv–RBD docking: The ClusPro web server (https://cluspro.bu.edu/ publications.php) was utilized for conducting protein-protein docking, given its position as one of the top 10 protein docking servers. This server uses a specially crafted pairwise interaction potential designed for antibody-antigen complexes, following the principles of statistical mechanics and the "inverse Boltzmann" principle 24. Additionally, it uses a pairwise interaction potential known as Decoys as the Reference State (DARS), which generates an extensive set of docked conformations that exhibit strong shape complementarity, irrespective of atom types, and relies on the interaction frequency derived from these DARS conformations. Consequently, it integrated DARS into the energy function employed in their docking program, PIPER 25. Initially, the SwissModel web server (https://swissmodel.expasy.org/) was utilized to construct the 3D model of isolated scFv, and subsequently, a Ramachandran plot (https://swift.cmbi.umcn.nl/ser-vers/html/ramaplot.html) was generated to validate its structural conformity. To prepare the input for the docking analysis, the modeled "scFv" was used as the receptor, and the following PDB IDs were used as antigens: 8WRL 26, 7ZF7 27, 7EKF 28, 8DF5 29, 8BCZ 30, 7EKC 28, 7TN0 31, and 7CH5 32. These correspond to the XBB.1.5 and BA.2 lineages, which are currently circulating VOIs, as well as the alpha, beta, delta, gamma, and omicron variants (previously known as variants of concern), and the wild type, respectively. To achieve this goal, the process was initiated by eliminating all water molecules from all structures. Subsequently, hydrogen atoms were introduced and energy minimization was performed using UCSF Chimera 33.
Results :
Construction of the recombinant PET28a vector: To produce a large amount of antigen for the biopanning process, the cDNA sequence of SARS-CoV-2 RBD protein was synthesized by RT-PCR and cloned into pET-28a vector. After purifying and inserting the RBD fragment into the PET28a vector, the recombinant construct was verified by PCR using RBD and vector-specific primers (Supplementary data 1). Finally, the RBD insertion into PET-28a were confirmed by sequencing of RBD fragment as follows:
>matched sequence
AGAGGTGATGAAGTCAGACAAATCGCTCCAGGGCAAACTGGAAAGATTGCTGATTATAATTATAAATTACCAGATGATTTTACAGGCTGCGTTATAGCTTGGAATTCTAACAATCTTGATTCTAAGGTTGGTGGTAATTATAATTACCTGTATAGATTGTTTAGGAAGTCTAATCTCAAACCTTTTGAGAGAGATATTTCAACTGAAATCTATCAGGCCGGTAGCACACCTTGTAATGGTGTTGAAGGTTTTAATTGTTACTTTCCTTTACAATCATACGGTTTCCAACCCACTAATGGTGTTGGTTACCAACCATACAGAGTAGTAGTACTTTCTTTTGAACTTCTACATGCACCAGCAACTGTT
Expression and purification of the receptor-binding domain (RBD): The recombinant vector was transformed into competent E. coli (BL21) and cultured in LB medium supplemented with kanamycin to select positive clones for induction. By adding 0.5 mM IPTG, the expression of RBD was induced. The recombinant RBD was purified on NI-NTA column, and refolded by dialysis. The SDS-PAGE results showed well integrity of the purified RBD protein and the total yield of the purified RBD was estimated to be about 1000 µg/ml (Figure 1).
RBD indirect ELISA: To assess the purity and integrity of RBD protein, an indirect ELISA using COVID-19 positive and negative serum samples was performed. Figure 2 shows the reactivity of COVID-19 sera to the purified RBD protein, indicating high integrity and purity of the recombinant RBD protein.
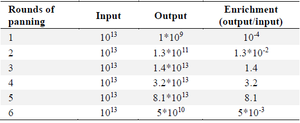
Analysis of Biopanning progress and Polyclonal phage ELISA: Six rounds of panning were performed using Tomlinson I Library with approximately 1013 CFU of phages on a 96 well ELISA plate coated with RBD protein. To increase the specificity of the selected phage, the RBD concentration was decreased gradually during the panning process (from 40 µg/ml in round 1 to 10 µg/ml in round 6). The enrichment of eluted RBD-specific phages was determined by phage quantitation following each round of selection. In the first round, the population of specific phage binders was low. However, after the second round, the titers of specific phages increased with the maximum enrichment observed in the fifth round (Table 3). A polyclonal phage ELISA was carried out to monitor binding affinity and specificity of RBD binders obtained from each round of panning. As illustrated in figure 3 the signals were remarkably low in the first round. However, the population of phage-scFvs gradually increased from the unpanned libraries towards fifth round. In the sixth round, the signal of diluted phages dramatically decreased, indicating the end of panning.
Identification of anti-RBD scFvs using monoclonal phage ELISA: A total of 100 colonies were randomly selected from the 5th round. Out of these, 20 clones were detected through ELISA. Among them, one clone with high reactivity with RBD (Figure 4) was selected for further analysis.
Sequencing of positive ScFvs: In order to verify the presence of full length scFvs, DNA was extracted from selected clones and used for PCR amplification using LMB3 and pHEN primers. Results indicated positive clones with the expected size (about 750 bp), which indicates the existence of both VL and VH fragments (Figure 5). Using http://www. imgt.org/3Dstructure-DB/cgi/DomainGapAlign.cgi, the CDR regions of the selected ScFv was determined as shown in table 4.
Evaluation of the reactivity of isolated scFv by ELISA: The pET-28a harboring the selected scFv sequence was transformed into competent E. coli BL21 (DE3) cells. The transformed bacteria were cultured and induced using 1 mM IPTG. The expressed scFv was purified by a Ni-NTA column. The expression and purity of the scFv were confirmed using SDS-PAGE analysis, which showed a molecular weight of approximately 25 KD (Figure 6). The specificity and reactivity of serially diluted scFv against RBD were assessed by indirect ELISA and results indicated that the selected scFv could detect the RBD protein with strong-binding (Figure 7).
scFv–RBDs docking analysis: The Ramachandran plot confirmed that the modeled scFv exhibited permissible phi and psi angles (Figure 8). The top-performing models have been chosen using the ClusPro algorithm, which employs a dedicated scoring function and clustering technique to evaluate the quality of receptor-ligand arrangements. Consequently, the mean scores for the best clusters were as follows for the wild type, XBB.1.5, BA.2, Alpha, Beta, Delta, Gamma, and Omicron variants: -264.4, -317.3, -357.3, -282.6, -264.1, -292.8, -332.5, and -268.3, respectively. According to the binding energy values, it is evident that the scFv and RBD in all models exhibit strong binding affinities. Additionally, the hydrogen bonds observed between the scFv and RBDs demonstrated robust interactions among the docked models (Table 5). From a comparative standpoint, even though all binding energies were elevated, it was evident that the wild-type, alpha, gamma, and BA.2 variants exhibited a greater abundance of robust interactions in contrast to the beta, delta, omicron, and XBB.1.5 variants. These results suggest that the discovered scFv has the potential to neutralize not only the wild-type virus but also the currently circulating VOIs and previous Variants of Concern (VOCs). All of the interactions are available in supplementary data 2, 3 and, 4.
Discussion :
Many researchers have attempted to find a cure for COVID-19 disease since its outbreak in early 2019. Monoclonal antibodies (mAbs) are approved as a versatile tool for treating infectious diseases, especially in high-risk patients 34. Specific human mAbs can be used as a safer alternative to attenuated or inactivated vaccines or immunoglobulin preparations for treating infectious diseases such as COVID-19 35,36. Recombinant mAb fragments, particularly single-chain variable fragment (scFv), have emerged as potential alternatives to mAbs. scFvs that can retain the specificity of whole mAbs are less expensive, and safer than mAbs. These stable fragments can be expressed in prokaryotic expression systems and engineered using phage display technology 35,37.
Therapeutic antibodies are able to target almost all proteins encoded by the SARS-CoV-2. Neutralizing antibodies (NAbs), in particular, can target the RBD of the spike glycoprotein. The RBD has been shown to be responsible for viral entry through binding to the ACE2 receptor in the human host and has become a promising target for NAbs 38,39. Currently approved mAbs, such as Etesevimab and Bamlanivimab, target the SARS-CoV-2 spike protein and have demonstrated efficacy in treating COVID-19. However, the continued emergence of viral variants, along with circulating mutations in the SARS-CoV-2 spike protein that can escape these approved mAbs, necessitates the exploration of new therapeutic antibodies 14. The present study focuses on isolating a novel human single-chain variable fragment antibody against SARS-CoV-2 RBD for potential therapeutic use. While neutralization activity has not yet been assessed, this scFv shows promise for further therapeutic exploration in future studies.
Prior research has indicated that the RBD is significantly exposed to the immune system, whereas the S2 segment remains hidden, resulting in its lower immunogenicity. Therefore, the S1 subunit of the S protein is considered a vital antigen for triggering immune responses. The efficacy of mAb that specifically target the RBD has been demonstrated in neutralizing SARS-CoV 40. In addition, immunization with the sequence of SARS-CoV-2 S protein generated neutralizing antibodies (NAbs) that mainly targeted the RBD 41. This implies that the removal of RBD-specific antibodies from SARS-CoV-2 patient serum may result in the loss of most neutralizing function. As a result, in this study, RBD recombinant protein was used as an antigen to identify novel RBD-specific scFv against SARS-CoV-2.
Here a strong-binding scfv mAb targeting the SARS-CoV-2 recombinant RBD using phage display technology is identified. For high expression yield of the RBD recombinant protein, RBD was expressed in E. coli expression system 42. Furthermore, to ensure that RBD maintained its native conformation a refolding system containing arginine, GSH and GSSG was used that was in accordance with He, Yunxia et al, who used the similar buffer for refolding of the recombinant RBD expressed in E. coli (BL21). The integrity and reactivity of the recombinant RBD was characterized using indirect ELISA and there was only a reaction between recombinant RBD and sera from COVID-19 patients, but not between recombinant RBD and sera from healthy non-infected individuals. Rahbar et al demonstrated that RBD expressed by E. coli and purified using Ni-NTA chromatography could react with antibodies present in the serum of immunized mice and individuals who had recovered from Covid-19 43. In the same context, Gao et al have indicated that the RBD sequence expressed in E. coli is capable of binding to the human ACE2 receptor and can be used for diagnosis and therapeutic purposes 44.
In this study, six rounds of biopanning were performed to eliminate non-specific clones along with the enrichment of specific clones. The results of the phage output titer in the screening rounds showed efficient enrichment towards an 80,000-fold increase in specificity against the S protein RBD. Among the six rounds of screening, finally a clone named 8-4 has correct fragments of light and heavy chains and promising-binding for RBD was identified from the fifth round. Phage display libraries have been successfully used to isolate scFv fragments against two previous Coronaviruses, SARS-CoV and MERS-CoV. In addition to binding to live viruses, these fragments exhibited a potent neutralizing effect through preventing viral entry 45-47.
In this regard, in recent years, several mAb fragments in scFv format against the RBD of SARS-CoV-2 virus were identified 48-50. Recent work by Delphine et al used phage display technology to isolate human scFv antibody fragments targeting the RBD of the SARS-CoV-2 spike protein. These scFvs exhibited binding to the spike protein of the SARS-CoV-2 Delta variant, although they did not bind to the Omicron variant. In comparison, present study also utilized phage display to isolate scFv antibodies against the SARS-CoV-2 RBD. Nevertheless, the robust binding of the present study scFv to the RBD observed in ELISA experiments suggests that candidates may hold similar therapeutic potential. Furthermore, according to the bioinformatics analysis, the present selected antibody is capable of recognizing the Wuhan strain as well as previous VOCs, VOIs, and VUMs of SARS-CoV-2.
In this study, bioinformatics tools were used to investigate the specificity and affinity of the discovered antibody against different SARS-CoV-2 variants. After confirming the modeled scFv by Ramachandran plot, the docking steps were performed using the Cluspro web server, and the antibody-antigen interactions were determined by LigPlot software. The use of these tools has been previously applied to the investigation of the interaction between Pseudomonas aeruginosa exotoxin A and a novel human scFv 21,51 as well as the interaction between the ESAT-6 antigen of Mycobacterium tuberculosis and two selected scFvs 52.
This study has several limitations that should be addressed in future research. First, while strong binding of the scFv to the SARS-CoV-2 RBD using ELISA was demonstrated, more rigorous binding affinity determination methods such as Surface Plasmon Resonance (SPR) or the Beatty assay was not conducted, to calculate the dissociation constant (kd) and kinetic parameters. Without these data, the precise affinity of the scFv remains undetermined. Second, although the scFv shows potential for therapeutic application, neutralization assays, such as RBD-ACE2 binding inhibition or live-virus neutralization assays, were not performed. These assays are critical for confirming the scFv’s ability to inhibit viral entry and validate its functional neutralization potential. Future studies will focus on addressing these limitations, conducting additional functional assays to validate the therapeutic potential of the scFv.
Conclusion :
In this study, a novel scFv with binding activity for the SARS-CoV-2 RBD, as demonstrated through ELISA binding assays was identified. Also, bioinformatics analysis confirmed that the antibody is effective against both the Wuhan strain, previous VOCs, VOIs and VUMs strains of SARS-CoV-2. While these results suggest potential neutralizing activity, further studies are necessary to confirm its functional efficacy.
Acknowledgement :
The authors would like to thank the Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran for providing research facilities and financial support toward the MSc thesis of the first author. Approval was granted by Research Ethics Committees of Vice-Chancellor in Research Affairs - Tabriz University of Medical Sciences (Date 2020-12-28/ID.IR.TBZMED.VCR.REC.1399.385).
Funding: This work was supported by Drug Applied Research Center (Grant numbers:66554).
Ethical approval :
This study was performed in line with the principles of the Declaration of Helsinki.
Conflict of Interest :
The authors have no relevant financial or non-financial interests to disclose.
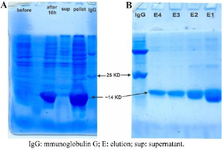
Figure 1. SDS-PAGE analysis of the expressed and purified RBD protein. A) The expression was induced by a concentration of 0.5 mM IPTG and the insoluble RBD had the expected size of about 14 Kda. Order of lanes from left to right: before induction, after 16 hr of induction, the amount of expression in soluble state, the amount of expression in insoluble state, and IgG marker. B) RBD protein was purified by Ni-NTA chromatography. Order of samples from right to left: Elusions E1, E2, E3, E4 and IgG as marker.
|
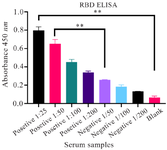
Figure 2. The results of ELISA for analyzing the reactivity of recombinant RBD and serum antibodies derived from both COVID-19 patients and healthy individuals. ELISA wells were coated with the 5 μg/ml of RBD, followed by blocking with 3% BSA. The results demonstrated a substantial binding of serum antibodies from infected individuals to the RBD antigen (OD=0.8), while the healthy samples exhibited a relatively lower reaction (OD=0.25). Thus, the integrity of the purified RBD was confirmed. The samples include 1:25 to 1:200 dilutions of positive (infected) samples and 1:50 to 1:200 dilutions of negative (healthy) samples. Error bars represent standard deviation. Statistical comparisons between groups were performed using a two-tailed t-test. (** p<0.01).
|
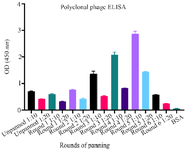
Figure 3. Analysis of the binding of phage clones to RBD from 6 cycles of biopanning using polyclonal ELISA. The results show the enrichment and presence of specific RBD phages in round 5. Therefore, single clones from this round were selected for further analysis. Table 3. The ratio of output to input (enrichment) for particular phage binders obtained from each cycle of panning
|
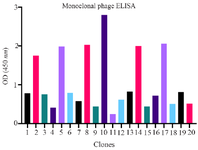
Figure 4. Monoclonal phage ELISA results. Upon analysis of phage clones selected from round 5 by monoclonal phage ELISA, clone number 10 exhibited a very strong reaction with RBD (OD=2.8). Therefore, it was selected for further evaluation.
|
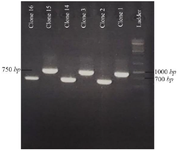
Figure 5. The results of PCR related to some full-length scFv fragments. scFvs with complete light and heavy chains showed a length of about 750 bp and were considered as positive clones.
|

Figure 6. scFv expression and purification analysis in E. coli BL21 (D3) strain. The expression induced by 0.5 mM IPTG. The scFv, with an estimated molecular weight of 25 KD, can be detected in the samples after induction, as well as in elution fractions 1 to 4. Order from left to right: IgG marker, flow-through, elution E1, E2, E3, E4, before induction of expression, 6 hr after induction and 16 hr after induction.
|
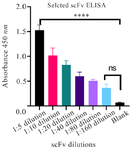
Figure 7. The specificity of scFv against RBD in different dilutions. The identified scFv revealed high reactivity with RBD (OD=1.527). Error bars represent standard deviation. Statistical significance was determined using one-way ANOVA followed by Dunnett's multiple comparisons test. A range of p-values from ****p<0.0001 (1:5,1:10 dilutions), ***p<0.001 (1:20 dilution), **p<0.01 (1:40,1:80 dilutions) indicate significant differences between the blank as control and dilutions (1:5-1:80). No significant differences were observed between blank and 1:160 dilutions.
|
Figure 8. Ramachandran Plot of scFv.
|
|

Supplementary data 1. The verification of the RBD sequence insertion into the vector. The amplified sequence by vector-specific primers (T7R-T7F) was heavier than amplified sequence using RBD and vector primers (T7R-RBDF and T7F-RBDR) and RBD-specific primers (RBDF-RBDR).
|
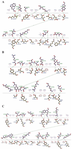
Supplementary data 2. Important interactions between scFv and wild, alpha and beta variants. A) Wild and scFv. B) Alpha and scFv. C) Gamma and scFv.
|
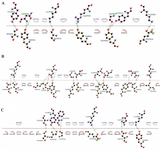
Supplementary data 3. Important interactions between scFv and beta, delta and omicron variants. A) Beta and scFv. B) Delta and scFv. C) Omicron and scFv.
|

Supplementary data 4. Important interactions between scFv and currently circulating variant of interest. A) BA.2 and scFv. B) XBB.1.5 and scFv.
|
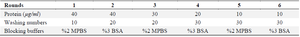
Table 1. The concentration of RBD, the frequency of washing, and the type of blocking buffers used in each biopanning round
BSA: Bovine Serum Albumin; MPBS: Marvel Skimmed Milk Powder in PBS.
|

Table 2. Primer sequences for VH and Vκ fragments
|
|
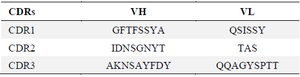
Table 4. The CDR regions of heavy and light chains variable regions of the discovered scFv
|
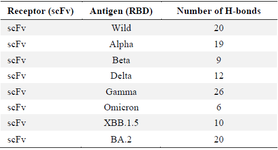
Table 5. Details regarding hydrogen bonds in all docking models
|
|