The Role of Disulfide Bridges in the Interaction of E. coli -Derived Recombinant SCARB2 and EV-A71's Capsid
-
Vo-Nguyen , Hai-Vy
-
Department Molecular and Environmental Biotechnology, Faculty of Biology and Biotechnology, University of Science, Ho Chi Minh City, Vietnam
-
Laboratory of Biosensors, Faculty of Biology and Biotechnology, University of Science, Ho Chi Minh City, Vietnam
-
Laboratory of Molecular Biotechnology, University of Science, Ho Chi Minh City, Vietnam
-
Vietnam National University, Ho Chi Minh City, Vietnam
-
Linh Thuoc, Tran
-
Department Molecular and Environmental Biotechnology, Faculty of Biology and Biotechnology, University of Science, Ho Chi Minh City, Vietnam
-
Laboratory of Biosensors, Faculty of Biology and Biotechnology, University of Science, Ho Chi Minh City, Vietnam
-
Laboratory of Molecular Biotechnology, University of Science, Ho Chi Minh City, Vietnam
-
Vietnam National University, Ho Chi Minh City, Vietnam
-
 Tran-Van, Hieu
Department Molecular and Environmental Biotechnology, Faculty of Biology and Biotechnology, University of Science, Ho Chi Minh City, Vietnam, Tel: +84 28 62884499; Fax: +84 28 38350096; E-mail: tvhieu@hcmus.edu.vn
Tran-Van, Hieu
Department Molecular and Environmental Biotechnology, Faculty of Biology and Biotechnology, University of Science, Ho Chi Minh City, Vietnam, Tel: +84 28 62884499; Fax: +84 28 38350096; E-mail: tvhieu@hcmus.edu.vn
-
Laboratory of Biosensors, Faculty of Biology and Biotechnology, University of Science, Ho Chi Minh City, Vietnam
-
Laboratory of Molecular Biotechnology, University of Science, Ho Chi Minh City, Vietnam
-
Vietnam National University, Ho Chi Minh City, Vietnam
Abstract: Background: Hand, Foot, and Mouth disease is an acute infectious disease caused by a group of enteroviruses, including Coxsackievirus A16 and Enterovirus 71. EV-A71-causing disease can give rise to severe complications in children, leading to meningitis, encephalitis, and even death. A potential approach to prevent the virus spread is inhibiting the invasion of EV-A71 into target cells, thereby helping to prevent not only the spread of EV-A71 in the community but also lessen the risk of outbreaks. EV-A71 cell’s receptor, human scavenger receptor class B member 2, SCARB2, was used as a trap to gather the virus and limit its spreading.
Methods: In this study, the recombinant receptor was expressed using Escherichia coli (E. coli) system. SCARB2 proteins expressed from E. coli BL21(DE3), and E. coli SHuffle® T7 Express were in inclusion bodies and subsequently refolded into soluble forms with recovery efficiencies of 57.57, and 82.2%, respectively. The presence of intramolecular disulfide bridges in the refolded SCARB2 was examined by SDS-PAGE in combination with Dithiothreitol (DTT). The two proteins were then utilized to evaluate the interaction with EV-A71 capsid by Indirect Enzyme-Linked Immunosorbent Assay (ELISA) at different pH levels to compare the adhesion efficiency.
Results: The results showed that SCARB2 protein expressed from E. coli SHuffle® T7 Express with disulfide bond modifications had better adhesion to the viral capsid. Notably, when the medium pH was lowered to acidic levels, the binding efficiency of recombinant receptors to the viral capsid increased.
Conclusion: To our knowledge, this study reported for the first time the activity of SCARB2 under extreme pH conditions and also revealed the crucial role of disulfide bridges in the interaction with EV-A71’s capsid. This finding contributed to the strategy using recombinant SCARB2 as a viral trap.
Introduction :
Infectious diseases in children have always been one of the biggest concerns of every country's health care system. Hand, Foot, and Mouth disease (HFMD) is a highly contagious disease with a viral causative agent, possessing common and easy ways of infection, leading to rapid spread and difficulty in control, putting burdens on the health care system and society. This disease mainly affects children under ten, especially infants and those under five, however, it still poses a threat to adolescents and occasionally to adults 1. The primary causative agent of HFMD is a group of enteroviruses from the Picornaviridae family, including Coxsackievirus A16 (CVA16), and Enterovirus 71 (EV-A71), and other CVAs including A7, A14 2. Once infected by EV-A71, the symptoms can appear from mild to severe, or even death 3. EV-A71 is a non-enveloped virus, with the capsid composed of four viral proteins (VP), VP1, VP2, VP3, and VP4, forming a twenty-sided structure with a diameter of 20-30 nm 4. VP1, VP2, and VP3 are exposed on the capsid surface, so they are tolerant to low pH, and easily detected and responded by the host immune system. In particular, VP1 is the target of neutralizing antibodies, putting VP1 under the selective pressure of the immune system, causing evolutionary changes in the capsid region and leading to the phylogenetic generation of new serotypes 5. Vaccination is always the most prioritized strategy in preventing viral diseases, however, this approach has been facing many unsolved challenges, including their efficacy and safety in long duration for a wide range of children ages, as well as the evolution of new species, 5-7. Another important factor, stated by the binding of antibodies to the viral capsid, as the rather blunt antibody was unable to penetrate into the conserved region to neutralize the virus 8. An alternative effort has been made to prevent the virus intrusion into host cells by utilizing its natural receptor, Scavenger Receptor Class B Member 2 (SCARB2) 9.
SCARB2 (also known as LGP85 or LIMP-2 or CD36b like-2) is a lysosomal type III transmembrane glycoprotein, belonging to the class B Scavenger receptor family, with a molecular mass of 55 kDa and composed of 478 amino acids, and is mostly localized to lysosomal membranes. SCARB2 is expressed in many cell types 10, and was declared to be the most important entrance for EV-A71 and other CVAs into host cells 2. The receptor has ten N-glycosyl sites, and two disulfide bridges (C274-C329 and C312-C318) 11. Many studies were performed to confirm the role of disulfide bridges in protein activity 12,13, raising the question of whether the two disulfide bridges in SCARB2 have any affection on their interaction with EV-A71.
Escherichia coli-derived recombinant SCARB2 demonstrated the ability to interact with EV-A71 in vitro 14. Nevertheless, the receptor was expressed in a non-disulfide bridge Escherichia coli (E. coli) strain. In this recent work, a professional-disulfide bridge-producing E. coli strain was utilized to generate another version of recombinant SCARB2. Then, the presence of disulfide bridges in the recombinant proteins after refolding was evaluated, and the role of the disulfide bridges in recombinant protein solubility and activity was investigated. Finally, the interaction evaluation was performed using an immune-related assay.
Materials and Methods :
Bacterial strains, plasmids, reagents and growth conditions: The recombinant vector pET-22b, and pET-SCARB2 carrying the encoding gene for SCARB2 receptor were provided by Laboratory of Biosensors, Faculty of Biology and Biotechnology, University of Science, Ho Chi Minh City, Vietnam 14. E. coli BL21(DE3) and E. coli SHuffle® T7 Express were recruited for recombinant protein expression. The two vector-carrying strains were cultured in Luria-Bertani (LB) medium containing 100 μg/ml of ampicillin, while E. coli BL21(DE3) grew optimally at 37°C, E. coli SHuffle® T7 Express was ideal at 30°C, according to the manufacturer's instructions (New England Biolabs). T7 promoter was employed as a protein expression regulator by using Isopropyl ß-D-1-thiogalacto-pyranoside (IPTG) (Biobasic). For band size identification, a protein marker (GE, 17044601), and a pre-stained protein marker (Thermo Scientific, 26612) were utilized. The recombinant receptor was expressed, refolded, and stored at -20°C 15.
SCARB2 expression in E. coli BL21(DE3) and E. coli SHuffle® T7 express: The expression of recombinant SCARB2 was performed as in the previous study 13 with some modifications for E. coli SHuffle® T7 Express. The vector pET-SCARB2 was transformed into competent E. coli BL21(DE3) and E. coli SHuffle® T7 Express strains. A single positive colony from each strain was selected for inoculation in an LB-ampicillin medium and grew at proper temperatures for each strain overnight. Then, the bacterial broth was sub-cultured at a ratio of 1:10 (v/v) into new LB media, and the expression was induced by the addition of 0.1 mM IPTG when OD600 reached 0.8-1.0. Following the induction, the cultures were maintained at proper temperatures (37°C for E. coli BL21(DE3), and 30°C for E. coli SHuffle® T7 Express) for 4 hr. Centrifuged cells were sonicated on ice in Phosphate-Buffered Saline (PBS) buffer (pH=7.4) to collect protein in total, soluble, and insoluble phases. Negative controls were performed with induced pET-22b-carrying E. coli and non-induced pET-SCARB2-carrying E. coli. To assess protein expression, Sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (using Tris-Glycine buffer with 12.5% gel in Bio-Rad electrophoresis systems) and Coomassie Brilliant Blue staining were conducted, followed by Western blot (using Cytiva blotting systems) with anti-SCARB2-Horseradish Peroxidase (HRP) antibody (Santa Cruz). The membrane plot was evolved with a proper amount of ready-to-use 3, 3', 5, 5'-Tetra methyl benzidine (TMB) (Thermo Scientific, 37574) and incubated at room temperature till the bands appeared. PBS buffer was used to halt the colorized reaction and the result was digitally recorded.
SCARB2 inclusion bodies refolding: In this research, two refolding strategies were separately utilized for two E. coli strains-expressed SCARB2, respectively. The strategy employed for E. coli BL21(DE3)-expressed SCARB2 was meticulously described 14. Accompanying, the strategy for E. coli SHuffle® T7 Express-expressed SCARB2 was carried out as previous work with some alterations 16. Harvested cells were on-ice-sonicated in cold extraction buffer [(50 mM Tris-HCl pH=8.0, 300 mM NaCl, 10% glycerol, 10 mM β-mercaptoethanol (βME), and 1 mM Phenylmethylsulfonyl fluoride (PMSF)]. The inclusion bodies were then separated by 13,000 rpm-in-20 min of centrifugation and followed by re-suspended along with shaking in extraction buffer containing 1% Triton X-100 for 30 min. The obtained precipitants were carefully rinsed five times in washing buffer (50 mM Tris-HCl pH=8.0, 500 mM NaCl). Subsequently, inclusion bodies were re-suspended in dissolving buffer (50 mM Tris-HCl pH=8.0, 300 mM NaCl, 20% glycerol, 10 mM βME, and 8 M Urea) overnight for collecting the solution containing solubilized recombinant protein. The mixture was then centrifuged at 13,000 rpm for 20 min to detach insoluble components. Eventually, the refolding products were evaluated through SDS-PAGE along with silver staining, and Western blot with anti-SCARB2-HRP antibody (Santa Cruz). The membrane plot was evolved with a proper amount of ready-to-use TMB (Thermo Scientific, 37574) and incubated at room temperature till the bands appeared. PBS buffer was used to halt the colorized reaction and the result was digitally recorded.
Dialysis was performed to remove refolding buffers from the aqueous phase containing solubilized SCARB2 after eliminating precipitants using PBS buffer (pH=7.4) with the reduction of Urea concentration from high, medium, to low concentrations (PBS 1× with 4 M Urea, PBS 1× with 2 M Urea, and PBS 1×), as the components in the buffers could impede subsequent experiments. To concentrate dialyzed protein, a 10 MWCO Amicon® Ultra centrifugal filter (Thermo Fisher) was utilized. The solubilized SCARB2 yield was ascertained by Bradford assay (Sigma).
Disulfide bridges confirmation: In this study, two E. coli strains were utilized for protein expression. Recombinant SCARB2 obtained from two E. coli-expressed strains were evaluated for the presence of disulfide bridges after refolding by the following method. The non-heated protein samples from two E. coli-expressed strains with and without DTT as a reducing agent were prepared for SDS-PAGE and subsequent with Coomassie Brilliant Blue staining. The protein sample containing disulfide bridges would migrate farther on the polyacrylamide gel than the lacking ones.
ELISA evaluation: 50% tissue culture infectious dose (TCID50) of EV-A71 and UV-inactivation preparation were conducted as in previous works 17-19. In short, a Petri dish containing dispersed EV-A71 was exposed to UV light for 30 min or longer in a Biosafety Cabinet at ambient temperature. Interaction assessment of recombinant SCARB2 with EV-A71’s capsid was demonstrated by the indirect ELISA as described with some adjustments 13. Firstly, 105 UV-inactivated EV-A71 virions in 100 µl carbonate buffer (pH=9.6) were applied to 96-well microplates, and incubation was performed overnight at 4°C. The coated plates were then fully blocked with 3% skim milk dissolved in Phosphate Buffered Saline-0.05% Tween (PBS-T) for an hour at room temperature. After three washes with PBS-T, recombinant SCARB2 with concentrations at 107, 108, 109, 1010, 1011, and 1012 molecules (diluted in PBS pH= 1.5, 3.5, 5.5, and 7.4) in 100 μl (of each concentration) were introduced to the plates, and incubation was performed for two hours at room temperature along with gentle shaking. The mixture was eliminated and followed by the addition of anti-His-tag-HRP antibody (Proteintech) at 1/20,000 dilution in 100 μl/well, continued with gentle shaking for one hour more. After discarding the antibody solutions and careful washes, the enzyme-linked reaction was performed with 100 µL of TMB (Sigma, T0440) at room temperature. Within 30 min, 100 μl of HCl 2 N was added to halt the reaction, the absorbance was assessed at 450 nm using an ELISA reader (Thermo). Negative controls were generated without the addition of virions coating or SCARB2 incubating. The results that were higher than the cut-off threshold created by the sum of negative control value and three times SD were labelled as positive. The data and plots were created and analyzed by GraphPad Prism using one-way ANOVA with multiple comparisons.
Results :
Recombinant SCARB2 expression: After transforming pET-SCARB2 into two E. coli strains, positive colonies were chosen for induction by IPTG to create recombinant SCARB2. Protein expression was confirmed by SDS-PAGE and developed with Coomassie blue. In both strains, overexpression of protein bands at about 45 kDa was observed (lane 3, 5; Figure 1A, C), which was the precise size for SCARB2 as predicted. Additionally, no overexpression bands with the appropriate size appeared in the negative control (lane 1, 2, Figure 1A, C), indicating that the E. coli itself did not possess the sequence encoding the target protein, also there was no leaky expression when IPTG was absent. Specific anti-SCARB2-HRP antibody was utilized for target protein detection through Western blot. The only bands visible on the blot proved that SCARB2 was the exact protein that was significantly expressed in the polyacrylamide gel, and the majority of the proteins were in the insoluble phase (Figure 1B, D). In conclusion, recombinant SCARB2 was synthesized in both E. coli BL21(DE3) and E. coli SHuffle® T7 Express as expected.
SCARB2 inclusion bodies solubilization and refolding: SCARB2 expression was performed in two pET-SCARB2 vector-carrying E. coli strains to obtain inclusion bodies. The rapid dilution method was utilized for SCARB2 obtained from E. coli BL21(DE3)/pET-SCARB2 in previous work 14. Briefly, harvested inclusion bodies proceeded for cleaning, dissolving, and refolding, then the final supernatant containing soluble SCARB2 was dialyzed and concentrated by 10 MWCO. Analyzing from the polyacrylamide gel (Figure 2A), except for lane 4, the targeted bands appeared in lanes 1-7 at 45 kDa, corresponding to the predicted molecular weight of SCARB2. A protein band in lane 2 showed that the dissolution of the inclusions was not complete, but a large amount of the target protein was successfully dissolved (lane 3). However, the refolding step was fully completed, as the protein band disappeared in lane 4. The refolding results were confirmed by the appearance of protein bands on Western blot probed with a specific antibody (Figure 2B). After dialyzing, the refolded protein remained in the soluble phase with a recovery efficiency of about 58% (Figure 2A, Table 1).
Simultaneously, harvested inclusion bodies expressed from E. coli SHuffle® T7 Express/pET-SCARB2 were obtained by dialysis method. After the dissolution step, the supernatant containing soluble SCARB2 was dialyzed and concentrated by 10 MWCO. The electrophoresis results on polyacrylamide gel showed that the targeted bands appeared in lanes 1-5 at 45 kDa (Figure 3A), corresponding to the molecular weight of SCARB2. The inclusions’ dissolution was not fully completed due to the appearance of a protein band in lane 2. Nonetheless, a massive portion of the target protein was effectively dissolved (lane 3). The corresponding results of Western blot confirmed the success of this refolding procedure (Figure 3B). SCARB2 after dialyzing remained soluble with a recovery efficiency of about 82% (Figure 3A, Table 1).
The two refolding methods utilized in this study succeeded in solubilizing the inclusions. SDS-PAGE gel images were processed with Gel Analyzer software to determine the percentage of target proteins in inclusions, and to calculate the ratio of SCARB2 in inclusions. The results indicated that the dialysis method yielded a better amount of protein in soluble form than the rapid dilution method, with higher recovery efficiency (Table 1).
Confirmation of disulfide bridges in recombinant SCARB2 after refolding: The presence of intramolecular disulfide bridges in the refolded SCARB2 was examined by SDS-PAGE in combination with DTT as a reducing agent. As can be observed from the electrophoresis gel (Figure 4), the DTT-treated protein sample, and the non-DTT treated E. coli BL21(DE3)-expressed SCARB2 migrated with the same distance, and at the molecular weight of SCARB2 (Figure 4, lane 1, 2, 3). Meanwhile, the non-DTT treated E. coli SHuffle® T7 Express-expressed SCARB2 migrated the farthest (Figure 4, lane 4).
Recombinant SCARB2-EV-A71 interaction evaluation: The interaction between EV-A71’s capsid and recombinant SCARB2 at different pHs (1.5, 3.5, 5.5, 7.4) was assessed by indirect ELISA using molar concentration rather than mass concentration. UV-inactivated EV-A71 was coated to a 96-well plate, followed by the addition of recombinant SCARB2 at different molar concentrations and pH levels. Once the recombinant receptor captured the viral capsid, it could be retained on the plate, and the interaction would be detected by anti-6xHis-HRP antibody with the addition of TMB substrate. The adhesion was evaluated at 450 nm, and the higher the absorbance, the stronger the interaction was indicated.
The ELISA results showed that the recombinant SCARB2 interacted with EV-A71’s capsid in dose-dependent and varied at four pH levels. When pH became more acidic, the viral capture ability became stronger (Figure 5). Specifically, with SCARB2/E. coli BL21(DE3), the interaction measured at pH=1.5 was higher than that at pH=5.5 and pH=7.4, with 2-fold and 4.5-fold, respectively (at a dilution of 1011 molecules) (Figure 5A). The same result was obtained with SCARB2/E. coli SHuffle® T7 Express, at 1011 molecules, the interaction measured at pH=1.5 was higher than that at pH=5.5 and pH=7.4, with 2.25-fold and 3.4-fold, respectively (Figure 5B). Furthermore, at a lower pH, the viral capture ability at one dilution is better than that at a higher pH. For example, with SCARB2/E. coli BL21(DE3) at pH=7.4, the lowest interactive dilution was 109, however, at pH=1.5, the lowest interactive dilution was 107. Likewise, with SCARB2/E. coli SHuffle® T7 Express at pH=7.4, the lowest interactive dilution was 108, and at pH=1.5, the lowest interactive dilution was 106. Obviously, the low pH condition facilitated important conformational changes in SCARB2, increasing the interaction ability 100 times when the receptor was exposed to the viral capsid at pH=1.5 compared to pH=7.4.
Simultaneously, the role of SCARB2’s disulfide bridge in the interaction with EV-A71 was partly revealed. At pH=1.5 and 1011 molecules, the interaction of EV-A71 with SCARB2/E. coli SHuffle® T7 Express was about 2 times higher than that of SCARB2/E. coli BL21(DE3). Similarly, at pH=7.4 and 1011 molecules, the interaction of SCARB2/E. coli SHuffle® T7 Express was 2.3 times higher than that of SCARB2/E. coli BL21(DE3) (Figure 5). Also, when the lowest interactive dilution of SCARB2/E. coli BL21(DE3) at pH=1.5 was 107, the lowest interactive dilution of SCARB2/E. coli SHuffle® T7 Express reached 106. At other pHs, the lowest interactive dilution of SCARB2/E. coli SHuffle® T7 Express was about 10-100 times lower than SCARB2/E. coli BL21(DE3) (Figure 5).
Discussion :
The soluble form of recombinant SCARB2 was previously reported 14. However, since SCARB2 is a human-derived protein, it required complex post-translational modifications, including ten N-glycosylation sites along with two disulfide bridges, both of which may be involved in the correct folding of the receptor 11. Therefore, in this study, to evaluate the role of disulfide bridges in supporting correct folding, and increasing the solubility of the target protein, SCARB2 was expressed in both E. coli BL21(DE3), and E. coli SHuffle® T7 Express. Specifically, E. coli SHuffle® T7 Express was ideal for the expression of disulfide bridge-carrying recombinant proteins, with protein activity yielding several times higher than the previously popular E. coli Origami(DE3) 20. Unexpectedly, SDS-PAGE and Western blot analysis indicated that the recombinant SCARB2 receptor was only obtained in inclusion bodies when expressed in E. coli system. This demonstrated that disulfide bridges did not significantly contribute to the receptor's solubility. Besides, the role of disulfide bridges in the interaction with EV-A71 remained unclear, raising a question to investigate, and obtaining the soluble form of recombinant SCARB2 was required.
Inclusion bodies are aggregate and water-insoluble proteins, due to incorrect folding structure 21,22. Inclusions are formed in the cytoplasm of E. coli for two main reasons: the expression systems, and the nature of the proteins. The limitation of the host system is the most common issue, including overexpressed protein leading to machinery overload and inclusion bodies formation. In addition, the E. coli system lacks some necessary post-translational modifications for protein folding, such as disulfide bridge formation and glycosylation 22. This factor is also directly related to ensuring proper expression of target proteins, especially viral or mammalian proteins. As the inclusion body exists in the aggregation, leading to the incorrect folding and configuration of targeted proteins, also affecting its biological activity. Two refolding methods were carried out to dissolve the SCARB2 inclusions 14,16. Accordingly, the presence of EDTA and PMSF in the lysis solution inactivated metal-dependent proteases and serine proteases, inhibited proteolytic cleavage, and increased the yield of target proteins in inclusions 23. The cold centrifugation removed impurities, likely bacterial soluble proteins, cell debris, cell membranes, nucleic acids, etc., which can affect the refolding efficiency 23. A washing buffer containing Triton×-100 removed untargeted proteins and cell membranes, as it broke the hydrogen bonds in the phospholipid bilayer of the cell membrane by its polar head group in the molecule 24. The dissolving buffer contains Guanidine HCl, and Urea broke incorrect bonds in the protein molecule, in order to solubilize the inclusions. The presence of bleach at low concentrations (Tween 80) and osmotic agents (sucrose, glycerol) reduced protein aggregation and increased refolding efficiency 23. After refolding, the recovery efficiencies of the two proteins were relatively high (with 58% for SCARB2/E. coli BL21(DE3), and 82% for SCARB2/E. coli SHuffle® T7 Express). Along with that, based on the major bands that appeared on each blot after dialyzing (Figure 2, lane 7, and Figure 3, lane 5), the two proteins were of sufficient quality for the next interaction evaluation section.
The refolding results in Western blot for both methods raised an issue to be discussed. The protein bands in the supernatant phase after refolding (Figure 2B, lane 5), and in the supernatant phase after dissolving (Figure 3, lane 3) disappeared on the membrane blot. The two solutions from the two methods both contained urea. Urea solutions contain isocyanates, which can cause carbamylation of the free amino groups of polypeptides 23. Carbamylation has been shown to affect protein structure and function 25, possibly affecting antibody-protein interaction by covering protein epitopes, and inhibiting the binding of antibodies to proteins, leading to the absence of targeted bands. After removing denaturing substances by dialysis, the protein bands reappeared (Figure 2B, lane 6, 7 and Figure 3B, lane 4, 5).
The role of disulfide bridges in interaction with EV-A71’s capsid was investigated. The native SCARB2 protein possesses two intramolecular disulfide bridges, located at C274-C329 and C312-C318 11. E. coli SHuffle® T7 Express was selected for protein expression to enhance disulfide bridge formation. βME was an indispensable component in the dialysis method. Initially, βME will cleave disulfide bridges in inclusions, but this disruption is reversible 26. In particular, the presence of glycerol supports the correct formation of disulfide bridges 27,28, dissolves the inclusions, and ensures its activity. The difference in protein migration in the DTT treatments and the two refolding methods may be due to the re-formation of the intramolecular disulfide bridge in the E. coli SHuffle® T7 Express-expressed SCARB2 after refolding. Without the reducing agent presence, the intramolecular disulfide bridges were preserved, so the molecular configuration of the protein remains in folded form, which is more compact than the straightened form due to DTT treatment. This result also indirectly confirmed that the refolding process in the presence of βME as a reducing agent is suitable for the targeted protein, providing higher recovery efficiency as well as activity. SDS is a commonly used detergent in SDS-PAGE. The combination of SDS, DTT, and high temperature, breaks down the secondary and tertiary structure of proteins. However, sole SDS can only break non-covalent bonds in protein molecules, such as hydrophobic and hydrogen bonds 29. Therefore, utilizing SDS-PAGE in combination with a sample loading solution without SDS and DTT will not significantly affect the intramolecular disulfides of proteins.
The reasons for the differences between the pHs-induced-interactions were discussed thoroughly 14. Although pH=5.5 was stated to be the point that the receptor would have significant flexibility 30; however, in this study, two more acidic pHs were applied to identify the interaction efficiency. The ELISA evaluation showed that the more acidic the pH, the stronger the interaction performed. Otherwise, under extreme pH, recombinant SCARB2 still retained its viral capture ability. This finding shall consolidate the idea of utilizing recombinant SCARB2 as a viral trap for humans, in order to prevent the virus spread and decrease the severe complications from EV-A71 infection. Furthermore, the role of disulfide bridges was partly confirmed. Although disulfide bridges did not contribute to the solubility of the recombinant SCARB2, the signals from disulfide-bridge-carrying protein were significantly better than the lacking ones. The presence of disulfide bridges may improve the folding configuration of the protein, making it to be more similar to the natural one, and increasing the viral capture ability.
Nevertheless, since SCARB2 is a human-derived protein with complex post-translational modifications, aside from two intramolecular disulfide bridges, ten N-glycosyl sites could be the main factor affecting the dissolution of recombinant SCARB2. Simultaneously, the role of glycosylation in interacting with EV-A71 has not been elucidated 9,11, so further studies are required, with higher expression system employment. Vaccination is always the most priority preventive solution for children's diseases. However, regarding the challenges that have not been resolved, the novel findings from this study can give a valuable contribution to the development of EV-A71 preventive strategy. One of the potential approaches could be using recombinant SCARB2 as an extracellular trap to capture the virus to lessen the affection of EV-A71 during the HFMD seasons.
Conclusion :
In summary, recombinant SCARB2 was successfully expressed in both E. coli BL21(DE3) and E. coli SHuffle® T7 Express in inclusion bodies. Two refolding methods were performed to obtain the soluble form of SCARB2 with high recovery efficiency. To our knowledge, this study reported for the first time the activity of SCARB2 under extreme pH conditions and also revealed the crucial role of disulfide bridges in the interaction with EV-A71’s capsid. This finding contributed to the strategy using recombinant SCARB2 as a viral trap. In case only the disulfide bridges strengthened the interaction, not the glycosylation, a more sufficient Prokaryote system should be utilized for more convenience compared to higher expression systems. Over and above, novel findings from this work could encourage non-invasive EV-A71 preventive approaches other than vaccination.
Acknowledgement :
This research was funded by the Ministry of Science and Technology of Vietnam through a bilateral project between Austria and Vietnam under grant number NĐT.105.AT/23.
Conflict of Interest :
The authors declare that they have no conflicts of interest.
Compliance with ethical standards: This article does not contain any studies with human participants or animals performed by the authors.
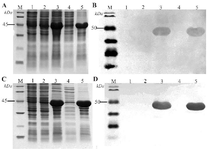
Figure 1. Expression of recombinant SCARB2 determined by SDS-PAGE with Coomassie brilliant blue staining (A, C) and confirmed by Western blot with anti-SCARB2-HRP (B, D) in E. coli BL21 (DE3) (A, B) and E. coli SHuffle® T7 Express (C, D). M, protein maker (A, C) and pre-stained protein marker (B, D); 1, E. coli/pET-22b (+IPTG); 2, E. coli/pET-SCARB2 (-IPTG); 3-5, E. coli/pET-SCARB2 (+IPTG); 3, total phase; 4, soluble phase; 5, insoluble phase.
|
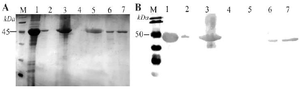
Figure 2. Refolding of SCARB2 inclusion bodies obtained from E. coli BL21(DE3)/pET-SCARB2 by rapid dilution method determined by SDS-PAGE with Silver staining (A) and verified by Western blot with anti-SCARB2-HRP (B). M, protein maker (A) and pre-stained protein marker (B); 1, inclusion bodies; 2, precipitated phase after dissolving; 3, supernatant phase after dissolving; 4, precipitated phase after refolding; 5, supernatant phase after refolding; 6, pre-cipitated phase after dialysing; 7, supernatant phase after dialysing.
|
Figure 3. Refolding of SCARB2 inclusion bodies from E. coli SHuffle® T7 Express/pET-SCARB2 by dialysis method determined by SDS-PAGE with Silver staining (A) and verified by Western blot with anti-SCARB2-HRP (B). M, protein maker (A) and pre-stained protein marker (B); 1, inclusion bodies; 2, precipitated phase after dissolving; 3, supernatant phase after dissolving; 4, precipitated phase after dialysing; 5, supernatant phase after dialysing.
|
Figure 4. Confirmation of disulfide bridges in recombinant SCARB2 after refolding. M, protein maker; 1, SCARB2/E. coli BL21(DE3) (+DTT); 2, SCARB2/E. coli SHuffle® T7 Express (+DTT); 3, SCARB2/E. coli BL21(DE3) (-DTT); 4, SCARB2/E. coli SHuffle® T7 Express (-DTT).
|
Figure 5. Evaluation of the interaction of recombinant SCARB2/E. coli BL21(DE3) (A), and SCARB2/E. coli SHuffle® T7 Express (B) with EV-A71’s capsid by indirect ELISA at different pHs.
|
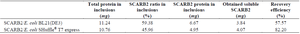
Table 1. Recovery efficiencies of the refolding processes
|
|