The Phenotypic and Molecular Identification of Phyllospheric Bacteria Possessing Antimicrobial Activity from Funtumia elastica (Preuss) Stapf.
-
 A. Adeniyi , Bolanle
Department of Pharmaceutical Microbiology, University of Ibadan, Nigeria, Tel: +234 802 3451480; E-mail: bolaadeniyi2022@gmail.com
A. Adeniyi , Bolanle
Department of Pharmaceutical Microbiology, University of Ibadan, Nigeria, Tel: +234 802 3451480; E-mail: bolaadeniyi2022@gmail.com
-
Ogunlana, Mercy
-
Department of Pharmaceutical Microbiology, University of Ibadan, Ibadan, Nigeria
-
B. Mahady , Gail
-
Department Pharmacy Practice, College of Pharmacy, University of Illinois, Chicago, United States of America
Abstract: Background: Unlike plant phytochemicals, little has been done to explore the metabolites from phyllosphere bacterial flora, some of which enabled them to survive interspecific competition through amensalism. This study evaluated the antimicrobial activity of metabolites from Phyllospheric Bacteria (PB) isolated from Funtumia elastica (FE), against selected bacterial and fungal pathogens. Phenotypic and molecular methods were used to identify the isolated phyllo-microbiota.
Methods: The PB were aseptically isolated by sonication. Their metabolites were obtained from the fresh overnight culture of the organisms. The cell-free supernatants containing the metabolites were used for antimicrobial assays against the pathogens. The DNA of the bacterial isolates were isolated using a NIMR-BIOTECH DNA extraction kit, while their 16S rRNA was amplified with the primer: 799F 5'-AACACGGATTA GATACC-3', 1193R 5'- ACGTCATCCCCACCTTCC-3', using SolisFast* Master Mix, (Solis Biodyne-Estonia). The BLAST of the sequence was done from the NCBI Genbank. The PB strains identified were submitted to NCBI and accession numbers were assigned to them.
Results: The phyllosphere of FE yielded 21 bacterial isolates: 7 Gram-positives and 14 Gram-negatives. The metabolites from these isolates showed varying degrees of bioactivity against Staphylococcus aureus (ATCC29213), Escherichia coli (ATCC 25922) Klebsiella pneumoniae (ATCC 35659); Trychophyton rubrum, Candida albicans and Microsporum canis. Fifteen bioactive isolates sequenced yielded four genera, Enterobacter (E. hormaechei 98.44%), Bacillus (B. cereus 100%), Pontoea (P. dispersa 99.72%), Staphylococcus (S. arlettae 99.72%).
Conclusion: Bacteria from FE phyllosphere, produced metabolites antagonistic (cidal) to some human pathogens. This has great potential for possible drug discovery.
Introduction :
In recent years, the search for and use of medicines and food supplements derived from plants has accelerated. Ethnopharmacologists, botanists, microbiologists, and natural product chemists are searching the earth for phytochemicals and "clues" that can be used to treat infectious diseases 1. Phytochemicals are biologically active chemical compounds that occur naturally, are found in plants and are beneficial for human health 2. Medicinal plants are rich in phytochemicals and can be optimized in their structure and processed into new drugs 3. Medicinal plants commonly used in our community can be an excellent source of these new drugs 4. The resistance of micro-organisms to classic antibiotics and their rapid development have aroused great attention in the treatment of infectious diseases 5.
Plant phyllosphere consist of aerial or above-ground parts of plants and are primarily one of the most prevalent microbial habitats on earth 6. The phyllosphere is dominated by leaves. The leaf surface habitat is very large with an estimated global leaf area of 508,630,100 km2 which means 1,017,260,200 km2 and implies a sufficiently large habitat for micro-organisms 7. The complex composition of phyllosphere microbes is influenced by some external factors such as light, UV radiation, atmospheric temperature, less water, and nutrients, which make them adapt to harsh environmental conditions 8.
Micro-organisms associated with plants may be endophytic or epiphytic depending on their location on the host plant. Endophytic organisms interact with and influence the internal part of plants, while epiphytic organisms interact and influence the exterior surface of plants. These organisms are not harmful to the plant but produce some useful substances that help in promoting the growth of the plant, providing resistance to pathogenic microbes and the production of secondary metabolites 9.
Bacteria are the most abundant micro-organisms found in the phyllosphere. It is estimated that 6.4×108 km2 of the global leaf surface harbours about 1026 bacteria, the most abundant colonizers of the phyllosphere 10. The culture method is widely used for the identification of the different microbes in phyllospheres. Thompson et al 11 identified 78 different bacterial species from sugar beet. However, this cultural method is possibly inaccurate in determining diversity. This is because of the presence of uncultured organisms. The use of culture-independent methods such as 16S rRNA sequences would give a more accurate and complete overview of the structure 8. Phenotypic characterization was based on comparing morphologic and phenotypic characteristics of type strains with that of the isolate to be identified. The 16S rRNA gene, being the highly conserved portion of the bacterial genome, is commonly used for taxonomic purposes in bacteria. The 16S rRNA is about 1550 bp long comprising of variable and conserved regions 12.
The average number of bacteria that are usually found on leaf surfaces is around 106-108 cells/cm. Proteobacteria, Firmicutes, Bacteroides, and Actinobacteria are the four major phyla that are generally found in the phyllosphere. Molecular studies have indicated that the highest bacterial inhabitants of the phyllosphere are firmicutes and alpha-, beta- and gamma-proteobacteria. Cyanobacteria, Acidobacteria, and Actinobacteria are also found to be frequently occurring in the phyllosphere. Methylobacterium and Sphingomonas are the most common genera of the class alpha-proteobacteria. Some of the factors that determine this bacterial assembly include the age of the plant, climatic conditions, the immune system of the plant, the species of plant and plant genotype and also, the soil type 8.
Environmental variables can change, while the geographical location of the plant and the plant genotype remain constant and stable. At times, the genotype has a major influence on the microbiome composition. However, geographical location has been reported to have the most significant impact. Some season-dependent microbial communities inhabiting the surface of perennial plants are almost the same from year to year, while others show significant variation with changing seasons 13.
Funtumia elastica (F. elastica) (Preuss) Stapf (Figure 1), is from the Apocynaceae family. Its common name is "Ireh" in Yoruba, and it is a forest tree growing in West and Central Africa 14. Funtumia elastica has a traditional history of ethnopharmacological use in the treatment of whooping cough, asthma, dysmenorrhea, wounds, and fungal infections. It is confirmed to exhibit significant antimicrobial activity against wide range of organisms such as Bacillus subtilis, Candida albicans (C. albicans), Pseudomonas aeruginosa, and Escherichia coli (E. coli), and diseases such as syphilis, gonorrhea, and hemorrhoids, amongst others 15. The stem latex of F. elastica is used for washing wounds, its leaves to treat hemorrhoids, and its bark powder in the treatment of respiratory ailments including asthma 14. It is also of great value in traditional medicine for the treatment and management of some diseases and disorders. A decoction from its bark is administered as a laxative and vermifuge. It is also included in prescriptions for problems associated with painful menstruation. The stem bark can be pounded and taken in spirits to cure hemorrhoids. The decoction from the stem bark is also used for treating chest infections like whooping cough. The decoction of its leaves is also used in treating mouth and venereal diseases 16. Despite the well-known antimicrobial activities of F. elastica, and its readily availability, there are no published studies on phyllosphere organisms on its leaf surface. This study therefore was designed to isolate, identify, and test their activity against clinical pathogenic bacterial and fungal isolates.
Materials and Methods :
Plant sample collection: The leaves of F. elastica were collected from the University of Ibadan Botanical Garden and aseptically transported in a sterile bag.
Isolation and identification of F. elastica phyllosphere bacteria: The plant leaves were aseptically collected into a pre-weighed sterile bag and then transported to the laboratory within an hour. The weight was determined. Eight grams (8 g) of the leaves were then transferred aseptically into a bottle containing 80 ml of sterile distilled water and aseptically sonicated for 10 min after which the sonicate was serially diluted into 10-1, 10-2, and 10-3 dilutions. The dilutions were inoculated into Tryptone Soy Agar (TSA) containing 0.8 ml of Nystatin in order to inhibit fungal growth. The inoculum was evenly distributed on the surface of the set agar using a sterile glass spreader. The plates were left on the bench for some minutes. They were then incubated on a transparent bench incubator, at room temperature for 4 days, close to the window for light accessibility.
Colony counts and subcultures from primary plates: The plates of the primary isolates were inspected and the colonies were counted. The morphologies of the various colonies were recorded. Isolated colonies were subcultured into fresh TSA plates without Nystatin. These plates were then incubated as described earlier. The results were then documented.
Biochemical tests on isolated organisms: Gram staining and biochemical tests were carried out and they include oxidase test, catalase test, and indole test. The growth of the organisms on differential media was also observed. These media include MacConkey agar, EMB, and MSA. The colour of colonies, texture, the appearance of colonies, and colour of plates were recorded.
Antimicrobial activity of cell-free supernatant (metabolites) of phyllospheric organisms: The antibacterial activity of the phyllospheric organisms against Staphylococcus aureus (S. aureus) (ATCC 29213), E. coli (ATCC 25922), and Klebsiella pneumoniae (K. pneumoniae) (ATCC 35659) was determined. The phyllospheric isolates were each cultured into 5 ml of sterile TSB and incubated for 48 hr. 20 ml of Muller Hinton agar (for bacteria), and Sabouraud Dextrose Agar (for fungi), were dispensed into MacCartney bottles and then sterilized. The molten agar was poured aseptically into sterile petri dishes and allowed to set. Pure colonies of the test organisms were resuspended in sterile distilled water, and standardized to 0.5 McFarland standard. A microbial lawn of the test organisms was made on the surface of the dried agar plates. A 6 mm diameter cork borer was used to bore wells on the plates. After 48 hr, of incubation of the phyllosphere isolates, about 2 ml of the broth culture of each organism, was dispensed into sterile cryovial tubes and centrifuged (Hospibrand-USA) at 1000 rpm for 5 min to obtain the cell-free supernatant. Sterile pasteur pipettes were used to dispense about 100 µl of the cell-free supernatant into the wells. Gentamicin (10 µg/ml) was used as the positive control for bacterial species, while Ketoconazole (10 µg/ml) for fungi. The plates were left for some minutes to allow for diffusion of the supernatant; after which they were incubated at 37°C for 24 hr, (for bacteria), and 25°C, for 48 hr (for fungi) 17,18.
DNA extraction and 16S ribosomal RNA amplification: About 5 to 6 colonies of each isolate were harvested into 100 µl of sterile molecular-grade water. The DNA from the cells was extracted by boiling in a water bath at 95℃ set at 10 min 19. This was then stored in a freezer at -20℃ awaiting Polymerase Chain Reaction (PCR).
Polymerase Chain Reaction (PCR): Two sets of primers were used to run the different PCRs; 799F and 1193R, the primer sequences are stated in table 1. The reaction was performed for 30 cycles in the PCR Thermal Cycler with initial denaturation at 95℃ for 2 min, final denaturation at 94℃ for 30 s, annealing of primers at 50℃ for 30 s, initial extension at 72℃ for 90 s and 10 min for final extension. The amplified product was visualized using an ultraviolet transilluminator. The nucleotide sequences of the 16S rRNA gene of 15 isolates were determined.
Results :
Total colony forming units for each dilution: The total colony forming units (CFU/ml) for the 10-1 and the 10-2 dilutions were 4.0×104 and 5.4×104, respectively.
Phenotypic identification of phyllosphere organisms: The physical characteristics of the phyllosphere organisms were determined by considering the form, elevation, appearance, pigmentation, and texture as shown in table 2. The cellular morphology of these organisms was also included in this table. The results of the biochemical test done on the isolates were recorded in table 3. This includes catalase, oxidase, and indole test.
16S ribosomal RNA amplification: Figure 2 shows the agar-gel electrophoresis result of the PCR amplification of the 16S rRNA of the bioactive isolates, while the isolated strains and their accession numbers are presented in table 4.
Antimicrobial activity of phyllosphere organisms: The result of the antimicrobial activity of the metabolites from phyllospheric organisms against the test pathogenic bacteria are recorded in table 5, while table 6, contains their antifungal activity.
Discussion :
The principle of interspecific antagonism amongst phyllospheric bacteria was applied in this study to determine the antimicrobial activity of metabolites obtained from bacteria isolated from F. elastica phyllospheres, against some selected human pathogens.
Twenty-one different colonies were isolated. These colonies observed primarily on TSA plates, ranged from white to cream to yellow and bright yellow in colour, indicating a mixed culture of different bacterial species. Of this number, Bacillus species were more prevalent, indicating their dominance of plant phyllosphere as reported by 20,21. Other Genera include Enterococcus sp, Staphylococcus sp, Ponteae sp, and Klebsiella sp, thus aligning our study with that of Ali M, et al 22 who reported similar genera in their study. The total colony forming units (CFU/ml) for the 10-1 and the 10-2 dilutions were 4.0×104 and 5.4×104, respectively, reflecting the reported numerical strength of bacteria in the phyllosphere 8.
There were 7 Gram-positives and 14 Gram-negative bacteria present, twelve of which were cocci in shape, while nine were rods (both long and short) (Tables 2 and 3). Earlier studies have shown a predominance of Gram-negative bacteria over Gram-positive ones on plant phyllosphere 23. It has been reported that quite a number of phyllospheric organisms are pigmented. This is advantageous to the phyllospheric bacteria as it helps to withstand the adverse effects of ultraviolet rays from the Sun 24,25. In a study conducted by Mazinani et al 21, 18 bacterial strains were isolated from Astragalus obtusifolius, 23 from Hippocrepis unisiliqousa, 32 from Prosopis juliflora and 31 from Xanthium strumarium. From F. elastica used in this study, 21 bacterial strains were isolated.
In this study, not all the phyllosphere bacteria`s metabolites showed bioactivity against the test pathogens. Isolate Fe-9b (BAT8), identified as Pantoea dispersa had broad-spectrum antimicrobial activity against Gram-positive bacteria, Gram-negative bacterium, and fungal. Pantoea dispersa W18, isolated from contaminated soil, was found to exert antimicrobial activity against Mycobacterium species, including Mycobacterium tuberculosis, an important human pathogen 26. Fe-24 which was eventually identified as Pantoea stewartii BAT21, showed activity against E. coli ATCC 25922, K. pneumoniae ATCC 35659, and C. albicans. Isolates Fe-7 (K. pneumonia BAT7), and Fe-10 (Enterobacter hormaechei BAT9), etc, were observed to show activity against at least one of the test bacteria. Based on this result (Table 5), 42% of isolated bacteria showed activity against at least one or more of the test pathogens. Lower activity against the dermatophytes was observed (Table 6). However, some of the isolates (33%) had activity against at least one of the dermatophytes tested. More of the phyllospheric bacteria showed activity against the bacterial pathogens than the fungi. This finding tends to agree with the trend in the study of Goryluk et al 20, who also reported low susceptibility of fungal pathogens to inhibitory effects of phyllospheric bacteria. In particular, the most active phyllospheric bacteria were Fe-9b (Pantoea dispersa BAT8) and Fe-24 (Pantoea stewartii BAT21). However, in the study by Mazinani et al 21, the most active strains were rods but Gram-positive.
Conclusion :
The result of this study shows that the isolated bacteria from F. elastica phyllosphere, produced metabolites that are antagonistic (cidal) to some human pathogens, including bacteria and fungi. Therefore, further research is necessary to purify, identify, and characterize these metabolites for possible drug discovery and development against these target pathogens.
Acknowledgement :
The authors wish to acknowledge the Technical Staff of the Department of Pharmaceutical Microbiology, University of Ibadan, Nigeria, for their technical input in this study.
Conflict of Interest :
All authors declared that there is no conflict of interest.

Figure 1. Funtumia elastica (Preuss) Stapf plant. (Photo by C. Delnatte).
|

Figure 2. Agar-gel electrophoresis result of the PCR amplification of the 16S rRNA of the bioactive isolates. Size of DNA marker used = 394 bp.
|

Table 1. Primers used for the amplification and sequencing of the 16S rRNA in this study
|
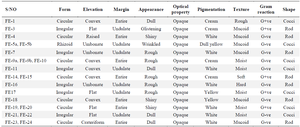
Table 2. The Gram reaction, cellular and colonial morphology of phyllospheric organisms of Funtumia elastic
|
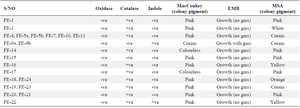
Table 3. Biochemical tests of isolated phyllosphere organisms
gms= green metallic sheen.
|
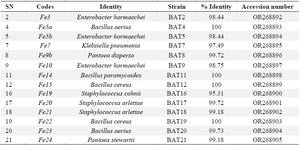
Table 4. The BLAST of the sequenced data and the assigned accession numbers
|
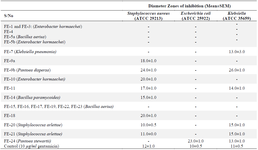
Table 5. Antibacterial activity of the metabolites from phyllosphere organisms against test organisms
SEM = Standard Error of the Mean.
|
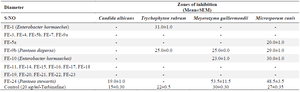
Table 6. Antifungal activity of metabolites from phyllosphere organisms against test organisms
SEM = Standard error of the mean.
|
|