Variation of ATM Gene Expression in Peripheral Blood Cells of Sporadic Breast Carcinomas in Iranian Patients
-
Foroughizadeh, Mohsen
-
Department of Medical Genetics, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran
-
 Mozdarani, Hossein
Department of Medical Genetics, School of Medical Sciences, Tarbiat Modares University, Tehran, P.O.Box: 14115-111, Iran, Tel: +98 21 82883830; Email: mozdarah@modares.ac.ir
Mozdarani, Hossein
Department of Medical Genetics, School of Medical Sciences, Tarbiat Modares University, Tehran, P.O.Box: 14115-111, Iran, Tel: +98 21 82883830; Email: mozdarah@modares.ac.ir
-
Department of Medical Genetics, Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran
-
Majidzadeh-A, Keyvan
-
Iranian Center for Breast Cancer, Academic Center for Culture, Education and Research (ACECR), Tehran, Iran
-
Kaviani, Ahmad
-
Faculty of Medicine, Tehran University of Medical Sciences (TUMS), Tehran, Iran
Abstract: The ataxia telangiectasia mutated gene (ATM), candidate for breast cancer susceptibility gene, encode a 350-kDa protein belongs to the core components of DNA-damage response machinery. Female AT carriers have at least 5-fold increase risk for breast cancer. Reduction in ATM expression is shown in multiple studies in breast tissues. We aimed to perform a research to measure the ATM mRNA expression in peripheral blood cells in breast cancer patients. Peripheral blood sample from 40 newly diagnosed, histologically confirmed female breast cancer patients was collected before surgery. Total RNA was isolated from blood cells using the RNX-Plus solution and reverse transcribed into cDNA. Real-time PCR was performed using the 2-∆∆CT method to calculate relative changes in gene expression by REST software. The Relative Quantitation (RQ) mean was 1.27 with the min. and max. equal to 0.20 and 3.34, respectively. Calculation of patient frequencies in different groups revealed that 17.5% had reduced expression lower than two fold decreases and 15% high expression more than two fold increases, but according to REST software there was no up-regulation or down-regulation compared to normal females. The findings of multiple studies consistent with this study indicate that the ATM gene may play an important role in breast cancer development and progression, and ATM expression is down-regulated in breast cancer tissues. Although, some of the results do not support a suppressor role for ATM in the development of sporadic breast cancer, 17.5% of our patients had under expression of ATM mRNA less than two fold relative to control.
Introduction :
Several genes are known to predispose women to breast cancer, which is a common disease with a complex etiology. Although mutations in BRCA1 and BRCA2 are recognized as risk factors for inherited breast cancer, somatic mutations in these genes are rare in sporadic breast cancers (1,2). In principle, a greater proportion of breast cancer cases within the population could be attributed to genes that are more frequently mutated but which may have a relatively low penetrance with respect to breast cancer (3,4).
The Ataxia Telangiectasia Mutated (ATM)gene is one candidate for such a susceptibility gene. The ATM a 350-kDa protein serine/threonine kinase, member of the phosphoinositide 3-kinase (PI3-kinase)-like family (PIKK) belongs to the core components of DNA-damage response machinery and acts as an intracellular sensor by recognizing double-strand breaks (DSBs). Numerous substrates involved in DNA repair are regulated by ATM protein kinase activity. ATM is the initiator of a signaling cascade that responds to DSB and thought to be master controllers of cell-cycle checkpoint signaling pathways (3,5,6).
Mutation of the ATM gene is the underlying cause of the rare autosomal recessive disorder Ataxia Telangiectasia (AT), characterized by clinical manifestations that include progressive cerebellar ataxia, skin, and ocular telangiectasia, immunological deficiency, neuronal degeneration, extreme cellular sensitivity to ionizing radiation (IR), premature ageing, hypogonadism, growth retardation, and predisposition to cancer. AT, described as a separate disease in 1957, occurs early in childhood with a frequency varying from 1 in 40,000 to 300,000 births in various ethnic groups (7,8).
The ATM gene was mapped to chromosome 11q22–23 in 1988 (9) and a single AT-mutated gene was identified in 1995 (2). More than 400 disease-causing mutations have been identified in ATM so far and about 70% of them result in premature termination of translation and truncation of the protein. The entire gene spans almost 150 kb of the genomic DNA, consists of 66 exons and is transcribed in a wide range of tissues to an mRNA of approximately 13 kb with a coding sequence of 9168 bp (5). An estimated 1.4% of the population carries a single ataxia-telangiectasia gene. Epidemiological studies on AT families have shown that AT heterozygotes also have an increased risk of developing cancer, in particular breast cancer, for which female ATM carriers have at least 5-fold increase risk compared with the general population. Seven percent or more of all breast cancer patients are likely to carry a single ataxia-telangiectasia gene. This gene appears to predispose carriers to breast cancer primarily with onset before age 65 or 70 years. Thus, the proportion of ataxia-telangiectasia heterozygotes among women with early breast cancer onset may be substantially greater than 7% (10,11), but its true magnitude is still uncertain (12).
Normal breast tissue shows a distinct pattern of ATM expression, the protein being found in the ductal epithelial cells but not in the surrounding myoepithelial cells. In contrast, in cases of sclerosing adenosis, a benign breast lesion, ATM is expressed in both the epithelial and myoepithelial cells. This up-regulation of ATM expression was associated with proliferation of the myoepithelial cells (13).
Several groups studied large cohorts of sporadic breast cancer patients and age-matched controls for nonsense or frame-shift mutations within the ATM gene. Neither group found evidence for a higher incidence of defective ATM alleles in the cancer patient cohort. Furthermore, the correlation between breast cancer and dominant negative forms of ATM arising from defined missense or intronic mutations within the ATM gene remained controversial. It seems that inactivation of the ATM gene by somatic mutations is not a common hallmark of breast cancer (5,14).
A role for the ATM gene in sporadic breast cancer is supported by many studies that have shown a LOH in the region of the ATM gene. This has been found in 40% of tumors studied. A causative association with the ATM gene has, however, been shown in only a few familial cases of breast cancer (3,15-17).
The epigenetic silencing of ATM expression occurred in locally advanced breast tumors suggesting a link between reduced ATM function and sporadic breast cancer (14).
HER2, Estrogen (ER) and Progesterone (PR) Receptors are currently used in routine patient care of breast cancer (18). Over-expression of human epidermal growth factor receptor type 2 (HER2), a 185-kD receptor occurs in 20 to 30% of invasive breast cancers. In general, tumors with a HER2 gene amplification have decreased overall survival and may respond to targeted therapy (19). ER positive tumors considered as lower penetrance alleles are thought to have characteristics of the luminal cell type and are frequently responsive to endocrine treatment. The PR is a prognostic marker, but the Oxford overview of adjuvant therapy does not support its ability to predict resistance to chemotherapy (20,21).
Multiple studies evaluated ATM gene expression in breast tissues, but so far, there was no available study on mRNA expression in tissues other than breast to determine a probable constitutional genetic predisposition factor in these patients. However, the expression of ATM gene in peripheral blood lymphocytes was evaluated in some cancers including bladder and lung (22,23). We aimed to perform a research to measure the ATM mRNA expression in peripheral blood cells in breast cancer patients and also their correlations in different groups.
Materials and Methods :
Study population: This study was approved by the local ethical committees. For the study, 40 newly diagnosed, histologically confirmed female breast cancer patients from Imam Khomeini hospital (Tehran, Iran) undergoing surgery were recruited between 2010 and 2011. There were no age, ethnicity, and tumor pathology or stage restrictions. Thirty three (83%) samples were invasive ductal carcinoma, 16 (48%) of these tumors were associated with an in situ ductal carcinoma. One sample was exclusively an in situ ductal carcinoma and 6 (15%) samples were other malignant breast tumors. None of the patients was treated with radiotherapy or chemotherapy before surgery. Twenty four control subjects with no prior history of cancer with the same gender for patients were recruited. The general characteristics of cancer patients are listed in table 1.
Blood sample collection: Peripheral blood sample from each participant was collected into EDTA tube before surgery and then transported immediately to the laboratory where the specimens were processed.
RNA extraction and cDNA synthesis: Total RNA was isolated from blood cells using the RNX-Plus solution for total RNA isolation (RNX-Plus solution; Cinnagen, CN. RN7713C) according to the manufacturer’s protocol. The kit utilizes the phenol chloroform method. Total RNA was eluted with 15 µl DEPC-treated water. One µl RNase inhibitor was added to prevent degradation of RNA (RiboLockTM RNase inhibitor; Fermentas Life Sciences). RNA concentration and purity were determined on a UV spectrophotometer by the 260 nm absorbance and 260/280 nm absorbance ratio, respectively. One µg of total RNA per sample was reverse transcribed into cDNA using the cDNA synthesis kit (QuantiTect Reverse Transcriptase kit; Qiagen, CN. 205311) according to the manufacturer’s protocol.
Quantitative real time PCR:The 2-∆∆CT method was used to calculate relative changes in gene expression determined from real-time quantitative PCR experiments. The probes and primers used for real-time RT–PCR were designed using the Primer Express software (version 2.0; Applied Biosystems) and synthesized by Takapou Zist Co. (Tehran, Iran). To avoid amplification of the contaminating residual genomic DNA, the probe and primer sets for each gene were designed around the junction region of two exons so that they were mRNA-specific. Human β actin gene was used as internal control to normalize input RNA amount, RNA quality and reverse transcription efficiency. The sequences of primers and probes are listed in table 2.
Real-time PCR was performed using the Applied Biosystems 7500 real-time PCR system according to the manufacturer’s protocol. Typical amplification mixtures (20 µl) contained the sample DNA (cDNA); 10 µl TaqMan® Universal PCR Master Mix (Roche Molecular Systems, Inc.; Part No. 4318157, New Jersey USA); forward and reverse primers and probes.
The efficiency of primers was 101.6 and 100.2 for ATM, and β actin, respectively. The thermal cycling conditions consisted of 1 cycle for 10 min at 95°C to activate the AmpliTaq Gold enzyme, and 45 cycles for 15 sec at 95°C and for 30 sec at 60°C. The quantitative PCRs were performed in duplicate for each sample, and the mean was used as the RQ value. The fold change in the ATM gene expression in patients (2-∆∆CT =RQ) normalized to β actin and relative to the expression at control samples was calculated for each sample where ∆∆CT = (CT,ATM - CT, β actin)Patient - (CT,ATM - CT, β actin)Control (24).
Statistical analysis: The Independent-Samples t-test or one-way ANOVA test was used, when appropriate to compare quantitative data. For evaluating relative expression of ATM mRNA in patients, REST (relative expression software tool) 2009 V2.0.13 was used. The statistical analysis was performed with the program SPSS 16.0 for windows.
Results :
Real-time RT-PCR of ATM mRNA expression in peripheral blood cells of patients showed the RQ mean equal to 1.27 (SD=0.76). The min. and max. was 0.20 and 3.34, respectively. Calculation of patient frequencies in different groups of RQ (Figure 1) revealed that 17.5% had under expression less than two fold decrease and 15% over expression more than two fold increase, but according to REST software there was no up-regulation or down-regulation compare to normal females. No correlation was found between ATM expression and tumor size (p=0.563), lymph node involvement (p=0.847), ER or PR (p=0.235 and p=0.605 respectively), or HER2 status (p=0.295). There was no significant correlation between RQ and different groups of ages, as well. Figure 2 shows amplification plot of some tumor and control samples.
Discussion :
The ATM protein kinase, encoded by the ATM tumor suppressor gene, is the apex of the repair pathway of DSB, a serious lesion occurring in DNA (5). ATM protects the integrity of the genome at different levels: (1) it mediates arrest of the cell cycle at G1/S, S, and G2/M to prevent the processing of damaged DNA; (2) it activates DNA-repair pathways; and (3) it induces apoptosis if the DNA damage is so detrimental that normal cell function can no longer be rescued (7).
The findings of multiple studies are consistent with the hypothesis that the ATM gene may play an important role in breast cancer development and progression, and also ATM expression is down-regulated in breast cancer tissues (20). Multiple studies revealed that the ATM mRNA level was significantly lower in breast cancer tissue than the adjacent normal tissue from the same patients or breast tissues from patients with benign breast diseases (25,26). This reduction was observed in breast tumors with or without LOH in the region of the ATM gene, suggesting that genetic events other than gene deletions could result in reduced ATM gene expression (27). However some studies showed that low expression of ATM in breast cancer tissue was related to a high rate of DNA mutation in cancer cells and a progressive cancer phenotype (28).
In some other studies ATM expression levels were the lowest in breast cancer tissue, compared to benign tumor tissue and normal breast tissue specimens (3,6,14). Reduced ATM expression was also associated with increased neovascularization in breast cancers, but a cause-effect relationship is yet to be demonstrated (6). Further, Vo et al found a highly significant correlation (p=0.0006) between reduced ATM mRNA abundance, as measured by real-time RT–PCR, and aberrant methylation of the ATM gene promoter. These findings indicate that epigenetic silencing of ATM expression occurs in locally advanced breast tumors, and establish a link at the molecular level between reduced ATM function and sporadic breast malignancy (14). But Luo et al have shown that in lymphocytes expressing ATM, the promoter region is completely demethylated. However, they were unable to correlate the methylation status and the variable ATM protein expression (5,29).
These results indicate that ATM down-regulation might be important at different levels and by different mechanisms in mammary carcinogenesis and that it may significantly contribute to the pathogenesis of breast carcinomas. Although, the results of Kovalev et al experiment do not support a suppressor role for ATM in the development of sporadic breast cancer (30).
ATM has more expression in higher grades of astrocytoma tumors (28), but in breast cancer patients high level of ATM expression in breast cancer tissue was linked to improved survival (25). ATM expression is a complex process, which is influenced by several reasons including DNA damage, accelerated protein degradation, mRNA instability, hyperphosphorylation of effectors proteins in the protein synthesis machinery (23).
Our results show no up- or down-regulation of ATM mRNA in peripheral blood cells in breast cancer patients, but in some other cancers reduced expression was seen. Jianlin et al evaluated the ATM protein expression levels of lung cancer patients in peripheral blood lymphocytes, and displayed that the ATM protein expression levels of lung cancer patients were significantly lower than that of controls (23). Among bladder cancer subjects the expressions of ATM was significantly lower in cases than in controls (p=0.04). A few studies showed that mRNA expression level of DNA repair or methylation-related gene in peripheral blood lymphocytes is associated with altered cancer risk. For example, reduced expression of mismatch repair genes and nucleotide excision repair genes confers significantly increased risk for head and neck cancer (22). However, our data correlate with Ye et al (2007) that no significant association was observed between ATM expression level and TNM stage (25).
ATM under expression was more observed in triple-negative breast tumors ER(-), PR(-) and HER2(-) (31). Furthermore, in another study the non-BRCA1/2 tumors with reduced ATM expression were more often ER negative, and PR negative (32), whereas according to our findings ATM expression in peripheral blood cells show no correlation in ER/PR status.
As shown in figure 1, 17.5% of patients had under expression of ATM mRNA less than two fold relative to control. The importance of this reduction should be evaluated in complementary studies that compare ATM expression in breast tissue and peripheral blood cells simultaneously.
Acknowledgement :
This study was financially supported by the Research Department of the Faculty of Medical Sciences, Tarbiat Modares University, Tehran, Iran. The authors would like to thank Mrs. Esmaeili for her technical help.
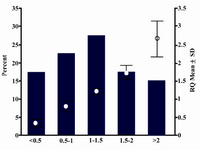
Figure 1. RQ mean and frequencies of patients in dif-ferent RQ groups.
|
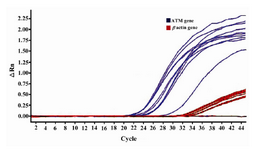
Figure 2. Amplification plot of some tumor and control samples
|

Table 1. Characteristics of patients with breast cancer
|

Table 2. Primer and probe sequences used in real-time PCR
|
|