In Silico Design and Selection of Anti-fungal AmB-polyene-analog Lead Molecules by Virtual Screening Method
-
Ferdosiyan, Marziyeh
-
Department of Biochemistry, Science and Research Branch, Islamic Azad University , Tehran, Iran
-
 Sardari, Soroush
Soroush Sardari, Ph.D., Drug Design and Bioinformatics Unit, Medical Biotechnology Department, Biotechnology Research Center, Pasteur Institute of Iran, Tehran, Iran, Tel: +98 21 66405535 Fax: +98 21 66465132 E-mail:ssardari@hotmail.com
Sardari, Soroush
Soroush Sardari, Ph.D., Drug Design and Bioinformatics Unit, Medical Biotechnology Department, Biotechnology Research Center, Pasteur Institute of Iran, Tehran, Iran, Tel: +98 21 66405535 Fax: +98 21 66465132 E-mail:ssardari@hotmail.com
-
Drug Design and Bioinformatics Unit, Medical Biotechnology Department, Biotechnology Research Center , Tehran, Iran
Abstract: A major group of drugs that have been approved for the therapy of systemic fungal infections are polyene antibiotics. Amphotericin B (AmB), one of the polyene antibiotics, has been used to treat serious systemic fungal infections by binding to sterols such as ergosterol in fungal cells membrane, and is believed to form pores in the membrane and create a transmembrane ion-channel. Since all eukaryotic cells contain sterols, using AmB can cause toxicity in mammalian cells; this is the most serious unwanted side effect. Therefore, there is still a need to develop suitable antifungal compounds to be entered in the drug development pipeline. In this study, we report the screening of various compounds from the Enhanced NCI database against ergosterol and cholesterol as receptors. The strategy employed is divided into two categories, screening and docking, respectively. Screening was performed using structure search based on AmB and molecular constraints to filter compounds with physico-chemical properties similar to the polyene macrolid antibiotics. The selected compounds were docked and scored to identify structurally novel ligands that make similar interactions to AmB. Our screening approach identified several molecules with high ranking criteria mentioned above. Among these compounds, two molecules, NSC 89270 and NSC 62792 were tested for their bioactivity against three fungal strains using broth microdilution assay that presented to have moderate antifungal activity against tested fungi. Thus, they could be possible lead compounds that grant further research on them to improve their potency and compare their mechanism of action in comparison to AmB.
Introduction :
Different kinds of mycoses, especially invasive, have increased dramatically in the past decades in relation to AIDS, cancer chemotherapy agents, transplant recipients and other immunosuppressed individuals (1 - 3). Part of the difficulty in treating fungal infection is the lack of a number of effective antifungal drugs (4). Marked differences were observed between mammalian membranes and fungal cell membranes. Mammalian membranes contain cholesterol and fungal cell membranes contain ergosterol as their principal steroid, whilst bacterial membranes do not normally contain steroids (5).
Biological action of polyen antifungal drugs is related to the cell membrane sterols
(6 - 8). Amphotericin B (AmB), one of the polyene macrolide antibiotics which have been acted by binding to sterols such as ergosterol in fungal cells membrane, and are believed to form hydrophilic pores in the membrane and create a transmembrane ion-channel (9, 10).
AmB is the most commonly used drug for many invasive or life-threatening mycoses. This is because it has the broadest spectrum of activity of any available antifungal agent and very rare induction fungal resistance. But, it is somewhat toxic, because it also has high affinity for cholesterol-containing memb-ranes, and its use in clinical treatment is limited to rather severe cases (11, 12).
While the azoles are much less toxic than amphotericin B, therapeutic failure and the emergence of azole resistance have limited their utility (13). The side effects of these drugs make the need for the necessity of new improved drugs. Hence, we try to find the new suitable analogues of AmB, by Virtual screening, which could be possible lead molecules with less toxicity. Traditional synthesis of a series of new compounds utilizing combinatorial chemistry and high-throughput screening can be carried out at high cost and also is time consuming. On the other hand, Virtual screening small molecule databases is now established as an effective paradigm for filtering compounds for drug discovery process (14, 15).
Virtual screening can apply docking and scoring of each compound from a dataset using a target receptor. Docking various ligands to target of interest followed by scoring to determine the affinity of binding and to reveal the strength of interaction has become increasingly important in the context of drug discovery. In general, it is important to understand the way the intermolecular interaction occurs by visualizing the docked positions of high-scoring compounds because many ligands are docked in different directions and may not fit in the manner of interactions that are known to be important for the target receptor (16, 17). Such studies become more difficult as the size of the dataset increases and CPU consumption and computer power become demanding.
Therefore, an alternative approach is to eliminate unpromising compounds before docking by restricting the dataset, by filtering the dataset based on similar features and appropriate properties. Using filters that reduce the number of possible interacting molecules can be highly effective in reducing the dataset to be docked (18, 19). In this study, we report screening a library of compounds from Enhanced NCI database against ergosterol, considered the ligand for AmB, by utilizing a fast, exhaustive docking application.
Materials and Methods :
Tools and materials used
For our present study, we used Enhanced NCI database, ChemAxon and Hex (version 5.1, 2008) servers. ChemAxon is developed to help chemists quickly and easily draw molecules, calculate physico-chemical properties based on structure, and it can convert name to structure and vice versa. Hex is an Interactive Molecular Graphics Program for calculating and displaying feasible docking modes of receptor-Ligand. Assuming the ligand is rigid, and it can superpose pairs of molecules using only knowledge of their 3D shapes (20). It uses Spherical Polar Fourier (SPF) correlations to accelerate the search for candidate low-energy conformation (21).
Enhanced NCI database is a Web-based service with different kinds of output features and links to other services for continued processing. Molecules in Enhanced NCI database are annotated by properties that include molecular weight, calculated LogP, number of hydrogen-bond donors, hydrogen-bond acceptors and number of rotatable bonds, Complexity, chiral centers, chiral double bonds (E/Z isomerism), net charge, drug likeness and PASS predictions. Biological activities of about 250,000 compounds in the Enhanced NCI Database are predicted by “PASS” prediction. This program predicts biological activity spectrum of compound based on its structural formula (22).
Screening
At first, AmB (Figure 1), one of the polyene macrolid antibiotics that is approved by FDA, was taken as an appropriate candidate to acquire new lead compounds based on structure-property relationship (SAR). Then, the Enhanced NCI database was screened for compounds similar to AmB structural features (structure based search) and by providing molecular constraints (property based search) according to properties of polyene macrolid antibiotics. The degree of similarity is dictated by the Tanimoto Index above 70% that given a numerical score for every compound to define the output ranking.
The physico-chemical properties such as log P value, H-bond donors, H-bond acceptors, topological polar surface and rotational bonds for about 60 polyene macrolid antibiotics were calculated using the ADME/ Tox filter server (23). Afterwards, using ChemAxon the structural of these obtained compounds that had less molecular complexity compared to AmB were sketched and converted to 3D structure. Receptors bound ligand AmB were docked to obtain the docked pose.
Therefore, the 3D structure of these compounds and receptors (ergosterol and cholesterol) complexes were identified via docking and their relative stabilities were evaluated using molecular dynamics and their binding affinities, using free energy simulations. The default parameters used for the docking process. Ultimately best compounds were selected to examine their antifungal activities.
Antifungal assays
For determination of MICs of the intended compounds, approved reference method M27-A has been published by the National Committee for Clinical Laboratory Standards (NCCLS) for testing with yeasts. And proposed reference method M38-P is being developed for filamentous fungi (24, 25). Two strains of yeast, Candida albicans (C. albicans) (ATCC10231) and Saccharomyces cervisiae (S. cervisiae) (PTCC 5052), one strain of filamentous Aspergillus niger (A. niger) (A420) were chosen to represent the effect of these intended compounds by using broth microdilution assay.
All strains were grown on Sabouraud glucose agar (SAB) growth media for 24 - 48 hr at 35 °C. Yeast inocula were prepared in sterile 0.9% NaCl solution and to make the inoculum of A. niger, the spores were dispersed in 0.1 % solution of Tween 20. The turbidity of the yeast and filamentous suspensions were adjusted respectively to 75 - 77% and 80 - 82% transmittance (T) by using a spectrophotometer at 530 nm. At last all inoculum suspensions were diluted (1:1000 for yeasts and 1:50 for filamentous) in Sabouraud maltose broth medium (SMB) to obtain the final test inoculum twice. All tested compou
Result :
We searched the Enhanced NCI database using structural features that are similar to AmB and then the results were restricted based on the computed properties ranges of the polyen macrolid antibiotics that are given in table 1.
The number of similarity identified hits was 500 and among those, about 200 compounds had less molecular complexity compared to AmB. We docked these 200 compounds using Hex and evaluated binding compatibility with the receptors based on docked energy (in kcal/ mol).
The docking tool generated 100 conformers for each docked molecule. By the virtual screening technique employed in this study, we identified specific diverse ligands that bind in a comparable manner to AmB binding for ergosterol and the nearest interaction energy values with ergosterol in comparison to AmB. The final screening based on dock and property filter approache identified ten compounds from the Enhanced NCI database for compatible binding with ergosterol. AmB on docking with ergosterol and cholesterol produced energy values of -170.9 and -186.0, respectively. Also, the “PASS” predicted for antifungal activity was about 0.889. The binding affinities between these compounds and ergosterol/ cholesterol and their physicochemical properties are tabulated in table 2. Their “PASS” predicted antifungal activities are also shown in table 2.
The structures of these compounds are presented in figure 2. Among them, NSC 89270 and NSC 62792 were novel molecules and available to purchase. Interaction of the NSC 89270 and AmB ligand molecules with the receptors is presented in figure 3. They were tested against mentioned fungal strains. Antifungal activities of the two compounds were compared with AmB and are shown in table 3. We reported the results against C. albicans (ATCC10231), S. cervisiae (PTCC 5052) and A. niger (A420) isolates, which they were semi-susceptible.
Discussion :
Virtual screening methods are routinely and extensively used to reduce cost and time in the drug discovery process. It has been clearly demonstrated that the approach utilized in this study is successful in finding novel antifungal lead compounds, based on structural and functional profile of AmB, from the Enhanced NCI database. The applied approach has the capacity to screen among compound for certain mechanism of action.
Conclusion :
Overall, our selection algorithm is efficient because speeds up the initial lead compounds identification. Further researches on these lead compounds provide an insight into discovery of stronger new antifungal compounds with better profile of activity. In future research work on properties of these compounds can be carried out in order to prove the effect of these compounds on the ergosterol present in the fungal cell membrane. In order to prove the effect of the compounds on the ergosterol present in the fungal cell membrane, the reversal agent such as ergosterol is to be tested for ability to affect the membrane dependant activity.
Acknowledgement :
The first author wishes to thank Drug Design and Bioinformatics Unit that all the study and experiments were carried out in it and for providing the supports.
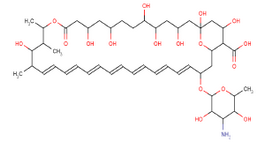
Figure 1. Structure of AmB
|
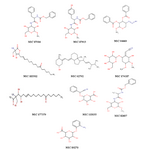
Figure 2. Compound structures were obtained from virtual screening
|
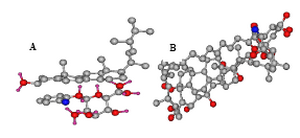
Figure 3. Interaction of compounds with ergosterol; A) NSC 89270-ergosterol and B) AmB-ergosterol
|

Table 1. Minimum to maximum ranges of about 60 polyene macrolid antibiotic properties
|
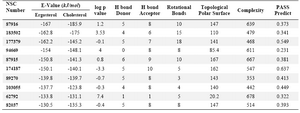
Table 2. Interaction parameters of the selected compounds with ergosterol and cholesterol and their predicted antifungal activities
|
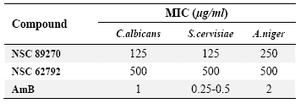
Table 3. In vitro activities of NSC 89270, NSC 62792 and amphotericin B against two strains of yeast and one strain of filamentous fungi after 48 hr
Highly-susceptible: MICs < 8 �g/ml
Susceptible: 8< MICs <100 �g/ml
Semi-susceptible: MICs 100
|
|