The Distinct Role of Small Heat Shock Protein 20 on HCV NS3 Expression in HEK-293T Cell Line
-
Basirnejad, Marzieh
-
Department of Molecular and Cellular Sciences, Faculty of Advanced Sciences & Technology, Pharmaceutical Sciences Branch, Islamic Azad University, Tehran, Iran
-
 Bolhassani, Azam
Department of Hepatitis and AIDS, Pasteur Institute of Iran, Tehran, Iran
Bolhassani, Azam
Department of Hepatitis and AIDS, Pasteur Institute of Iran, Tehran, Iran
-
Department of Hepatitis and AIDS, Pasteur Institute of Iran, Tehran, Iran
-
Sadat, Seyed Mehdi
-
Department of Hepatitis and AIDS, Pasteur Institute of Iran, Tehran, Iran
Abstract: Background: Hepatitis C (HCV) is known as a serious blood-borne disease that infects millions of people globally. NS3 is a conserved non-structural sequence of hepatitis C virus which has a major role in activating specific CTL responses. As known, there is no effective vaccine against HCV infection, thus it is required to design a specific regimen of vaccination. Recently, the strong immunological properties of Heat shock proteins (Hsps) led to their use as immunomodulators and an antigen carrier for subunit vaccine candidates. In the current study, the role of Hsp20 was evaluated as a HCV NS3 gene carrier in mammalian cell line.
Methods: At first, the recombinant plasmids of pEGFP-Hsp20, pEGFP-NS3, and pEGFP-Hsp20-NS3 were constructed and their accuracy was confirmed by digestion and sequencing. Then, all recombinant plasmids were transfected into HEK293T cells by Lipofectamine and TurboFect gene delivery systems. Finally, the expression of proteins was assessed by fluorescent microscopy, western blotting, and flow cytometry.
Results: In western blotting, the 47, 59, and 79 kDa bands were detected for pEGFP-Hsp20, pEGFP-NS3, and pEGFP-Hsp20-NS3, respectively. The percentage of NS3-Hsp20-GFP protein expression was ~67% by TurboFect and ~50% by Lipofectamine indicating high potency of TurboFect delivery system. Furthermore, the expression of Hsp20 (~83%) was higher than NS3 (~58%) in the cells transfected by TurboFect using flow cytometry analysis. This result was confirmed in the expression of Hsp20-NS3 fusion (~67%) in which Hsp20 increased the delivery of HCV NS3 in vitro. The same data were obtained by Lipofectamine transfection reagent.
Conclusion: Briefly, our data confirmed the role of Hsp20 as a suitable antigen carrier for DNA vaccine design.
Introduction :
Hepatitis C virus (HCV) is a single-stranded enveloped RNA virus with nearly 9600 nucleotides in length. Based on the RNA genome of the virus, hepatitis C is categorized into six genotypes in the world that the most common type is genotype 1. Eleven proteins are encoded by HCV genome including structural and Non-Structural (NS) proteins. NS3 is a highly conserved, non-structural protein containing a serine protease domain in N-terminal and a helicase/NTPase domain in C-terminal of protein 1. NS3 was suggested to be the best vaccine candidate for hepatitis C due to the induction of strong T-cell immune responses against HCV NS3 related to clearance of infection. However, no effective HCV vaccine has been found because of the genetic variability of host defenses and the potential of the virus to escape the host immunity 2,3.
Among different vaccine strategies, DNA vaccines have attracted a specific interest due to easy production, heat resistant, and safety. A number of evidences showed that CpG motifs in plasmid vectors stimulate B-cell activity and subsequently humoral immune system 4. The aim of vaccination is to provide long-term protection against infections. Due to the low penetration of plasmid DNAs into the cells, development of an effective adjuvant is necessary for designing DNA vaccines 5. Therefore, researchers are studying for proper combination of antigens with effective adjuvants or carrier molecules in subunit vaccines 6.
Recently, Heat shock proteins (Hsps) were proposed to increase immune responses against infectious diseases 7. Hsps were classified into different families based on their molecular weight 8,9. Among heat shock proteins, small HSPs are highly conserved proteins among all species which have a conservative α-crystalline domain (~90 amino acid residues). In addition, small HSPs have many functions such as protein folding, transportation, proteostasis, and immunity. Some small heat shock proteins are tissue specific in human such as HspB2, HspB3, α-crystalline (HspB4), HspB7, HspB9, and HspB10, while others are expressed in all human tissues including HspB1, αB-crystalline (HspB5), HspB6 (20 kDa) and HspB8 10-17. HSPs are capable of delivering antigens to major histocompatibility complexes (MHC) for stimulation of adaptive immunity 6,7,18,19.
In this study, the plasmid DNAs encoding Hsp20, NS3, Hsp20-NS3 were generated and their expression was evaluated in mammalian cell line using a cationic polymer (TurboFect) and a cationic lipid (Lipofectamine). TurboFect transfection reagent is a cationic polymer in water which forms compact, stable and positively charged complexes with DNA facilitating gene delivery into eukaryotic cells (www.thermofisher.com). The data indicated that TurboFect transfection reagent was more effective than Lipofectamine for delivering the recombinant DNAs in HEK-293 T cells. Also, Hsp20 enhanced the transfection and expression of HCV NS3 in vitro. The obtained data would be a basic step for immunological studies in future.
Materials and Methods :
Construction of the recombinant plasmids: The full length of Hsp20 sequence was synthesized in pQE30 by Biomatik Company. For generation of pEGFP-Hsp20, the Hsp20 gene (Accession No: NM_001012401) was digested by BamHI/HindIII and subcloned into BglII/HindIII sites of pEGFP-C1. The eukaryotic vector (pcDNA3.1) harboring the immuno-genic and conserved region of HCV subtype 1a NS3 gene (1095-1379 aa, No: EU781798.1) was digested by XhoI/HindIII (Thermo scientific Fastdigest) and sub-cloned into pEGFP-C3 expression vector. To prepare the pEGFP-Hsp20-NS3, at first, the NS3 gene was ligated in SalI/HindIII restriction sites of pQE-Hsp20 using T4 DNA ligase. Then, the fusion of Hsp20-NS3 was digested by BamHI/HindIII and subcloned into BglII/HindIII cloning sites of pEGFP-C1. The Escherichia coli (E. coli) DH5α strain was transformed by all the recombinant vectors. After the extraction of plasmids from single colonies using DNA extraction kit (Qiagen), they were confirmed by digestion and sequencing. The recombinant pEGFP-NS3, pEGFP-Hsp20, and pEGFP-Hsp 20-NS3 plasmids were provided in large scale using DNA extraction kit (Qiagen, Germany) and quantified by NanoDrop spectrophotometry. The purity of plasmids was determined as OD260/OD280 ratio. This ratio was ~1.85 for all plasmids indicating their purity. Figure 1 shows schematic representation of cloning process.
Cell culture: Human embryonic kidney cells (HEK-293T) were cultured in Dulbecco's Modified Eagle's Medium (DMEM, Gibco) supplemented with 10% Fetal Bovine Serum (FBS, Gibco) at 37oC in presence of 5% CO2 atmosphere. After several passages using trypsin EDTA, the proliferated cells were counted by trypan blue 1X with hemocytometer and divided into 6-well plate.
Transfection of plasmid DNAs into HEK-293T cells using Lipofectamine 2000: The day before transfection, the 5×105 cells were counted and seeded into 6-well plates. The optimal cell confluency for effective transfection was considered 70-80%. For generation of Lipofectamine-plasmid DNA complex, 150 μl of serum-free medium was mixed with 10 μl of Lipofectamine (Invitrogen) and incubated for 5 min at room temperature. Then, 150 μl of incomplete DMEM was mixed with 4 μg of each plasmid (i.e. pEGFP-NS3, pEGFP-Hsp20, pEGFP-Hsp20-NS3, and pEGFP-C1 as a positive control), added to Lipofectamine solution, mixed gently, and incubated for 30 min at room temperature. After that, transfection complexes were added to each well and the medium was replaced after 6 hr of incubation at 37oC with the complete DMEM containing 10% FBS and 1/100 penicillin/streptomycin.
Transfection of plasmid DNAs into HEK-293T cells using TurboFect: For generation of TurboFect-plasmid DNA complex, 7 μl of TurboFect (Thermo scientific) and 4 μg of plasmid were mixed and incubated for 15 min at room temperature. Finally, the complexes were added dropwise to each well in serum-free medium. Six hours after the cell transfection, the medium was replaced with the complete DMEM medium. After 48 hr, transfection efficiency using TurboFect and also Lipofect-amine was evaluated by fluorescent microscopy, flow cytometry, and western blotting. The non-transfected cells and the cells transfected by pEGFP-C1 were used as negative and positive controls, respectively.
Fluorescent microscopy and flow cytometry analysis: The quality and quantity of protein expression were monitored by GFP expression as a reporter gene using fluorescent microscopy (Envert Fluorescent Ceti, Korea) and Fluorescence-Activated Cell Sorting (FACS) caliber flow cytometer (Partec, Germany), respectively. For flow cytometry analysis, the cells were harvested by trypsin and the cell pellets were resuspended in 1 ml PBS (pH=7.4). Then, GFP expression in transfected cells was compared to non-transfected cells.
Western blot analysis: For western blotting, the cells were harvested by trypsin, and the cell pellets were resuspended in PBS. Total cellular proteins were solved in 6X sample buffer containing Tris-HCl (0.5 M), glycerol, SDS, and 2-Mercaptoethanol (2%). The samples were separated on 15% acrylamide gel and transferred to nitrocellulose membrane. The membrane was incubated in blocking buffer (TBS 10X, 0.1% Tween 20, BSA, Merck) and washed with TBS10X and 0.1% Tween 20. Then, anti-GFP polyclonal antibody conjugated with horseradish peroxidase (1:10000 v/v) was used to detect the proteins of interest in the presence of DAB substrate (Roche Diagnostics-Germany).
Statistical analysis: Statistical analysis (Student’s t-test) was performed by Prism 5.0 software (GraphPad, San Diego, California, USA) to analyze the percentage of NS3-GFP, Hsp20-GFP, and Hsp20-NS3-GFP expression using flow cytometry. The value of p<0.05 was considered statistically significant. Similar results were obtained in two independent experiments.
Results :
DNA constructs expressing NS3, Hsp20, and Hsp20-NS3 genes: The pEGFP plasmids are mammalian expression vectors containing GFP sequence and Kanamycin resistance marker which were selected in the current study. The recombinant plasmids were prepared by subcloning as mentioned in Methods section. The recombinant pEGFP-Hsp20 digested by restriction enzymes showed the clear bands of ~720 and ~581 bp related to GFP and Hsp20, respectively. The recombinant pEGFP-NS3 digested by enzymes indicated a ~861 bp band related to NS3. In addition, the recombinant pEGFP-Hsp20-NS3 cut by NheI/HindIII showed the clear bands of 720 and 1442 bp related to GFP and Hsp20-NS3, respectively as shown in figure 2.
Transient expression of proteins in HEK-293T cells: The transfection efficiency of TurboFect and Lipofectamine reagents was confirmed by fluorescent microscopy and flow cytometry as shown in figure 3. Flow cytometry analysis indicated that the expression of pEGFP-Hsp20, pEGFP-NS3, and pEGFP-Hsp20-NS3 proteins using TurboFect transfection system was significantly higher than Lipofectamine (p<0.05). These results were obtained using the percentage of protein expression using GFP reporter marker. The percentages of Hsp20-GFP, NS3-GFP, Hsp20-NS3-GFP, GFP expression were 83.10±2.24, 58.02±1.19, 67.23±1.12, 90.15±1.99 for TurboFect system, and 66.01±1.63, 43.31±0.85, 50.49±1.68, 86.45±1.75 for Lipofectamine, respectively. Indeed, Hsp20 could enhance NS3 DNA delivery in the cells significantly more than NS3 DNA alone using both TurboFect and Lipofectamine transfection reagents (Figure 4, p<0.05).
Western blotting: Western blot analysis indicated the successful expression of Hsp20-GFP, NS3-GFP, Hsp20-NS3-GFP, GFP proteins using anti-GFP antibody as shown in figure 5. The data indicated the clear bands of ~47, ~59, ~79 and ~27 kDa for Hsp20-GFP, NS3-GFP, Hsp20-NS3-GFP and GFP, respectively using DAB substrate.
Discussion :
The studies showed that HCV-specific CTL responses are very important for control of viral replication in chronically infected individuals 20. In 2001, Lazdina et al showed that NS3 elicits Th1 immune responses in a DNA vaccine significantly higher than that in a recombinant protein vaccine 21. In 1996, Missale et al examined the peripheral blood T cell proliferative responses against HCV core, E1, E2, NS3, NS4 and NS5 recombinant antigens. The results showed that 43% of T-cell responses were induced by NS3 antigen 22. The similar data were obtained by Tsai et al for a considerable CD4+ T-cell proliferation to NS3 in patients with acute hepatitis C 23.
In addition, the studies showed that NS3-specific immune responses are cross-reactive in various genotypes of HCV 24. Thus, NS3 was considered as a vaccine candidate. A major problem during development of DNA vaccines was their weak immunogenicity and the poor potency of such vaccines which may integrate into the host cell. For these reasons, there is a great necessity for the use of adjuvant to stimulate the T-cells 25.
In this study, HCV NS3 gene as an antigen candidate was effectively cloned in the eukaryotic expression vector alone or fused with Hsp20 as an effective carrier or adjuvant. The purity of plasmids was similar for using in transfection method. In general, the results showed that Hsp20 could increase the transfection efficiency of HCV NS3 in HEK-293T mammalian cells. There are some experiments for increasing the potency of HCV NS3 antigen. For instance, Naderi et al designated Interleukin-12 (IL-12) as an adjuvant for HCV NS3 DNA vaccine 26. Jiao et al showed that the cellular and humoral responses of the recombinant NS3 under the effect of CpG as an adjuvant was enhanced in animal model 27. Qazi et al used HSP70 as a suitable vaccine adjuvant. Based on this research, HSPs were selected as a carrier that conjugated to the malaria antigen EB200 and delivered both chimeric protein and DNA construct 28.
On the other hand, Barrios et al showed that mice immunized with peptides or oligosaccharides conjugated to the mycobacterial Hsp70 generated high titers of IgG antibodies in the absence of any adjuvant. Indeed, the use of Hsp as carrier in conjugated constructs for the induction of anti-peptide and anti-oligosaccharide antibodies could be of value in the design of novel vaccines 29. Ebrahimi and Tebianian also showed that the linkage of antigen with limited potency to an appropriate carrier such as C-terminal 28 kDa domain of mHSP70 (HSP70 359-610) containing an 18 kDa peptide binding region (aa 359-540) can increase its immunogenicity without any side effects 30. Moreover, Hsp20 contains α-crystalline domain known as a ligand for toll-like receptor 2/4 located on dendritic, macrophage, mast, monocyte, microglia, neutrophil, and T-cells 31.
Alvarez et al showed that the Leishmania Hsp20 as DNA vaccine is antigenic during natural infections. They indicated that 62% of the Leishmania infected animals, elicited considerable humoral responses against Hsp20 32. Ortiz et al reported that DNA fragment containing B and T cell epitopes of the N-terminal region of Hsp20 with other Babesia bovis antigens elicited high levels of specific IgG antibodies 33. Brown et al also reported that Hsp20 is a highly conserved protein between species and has B and T lymphocyte epitopes 34. Bepperling et al investigated bacterial small heat shock proteins such as Hsp17.7 and Hsp20.2 proteins. The results demonstrated that Hsp20.2 has strong chaperone activity as compared to Hsp17.7 35.
Regarding the roles of NS3 as an antigen candidate and Hsp20 as an antigen carrier and adjuvant, DNA constructs of pEGFP-NS3, pEGFP-Hsp20 and pEGFP-Hsp20-NS3 were prepared and their delivery was evaluated using TurboFect and Lipofectamine in mammalian cells. The researchers classified non-viral gene delivery systems in three groups: Naked DNA delivery, Lipid-based, and polymer-based delivery 36. Several studies used Lipofectamine and TurboFect as a vehicle to transport DNA constructs into the cells. In 1995, Lin et al transfected vHCV1027-1207 encoding the NS3 into BHK-21 cells with Lipofectamine 37. In 2003, He et al showed a suitable transfection of pRcHCNS3-5’ and pRcCMV expressing HCV NS3 into QSG7701 cells using lipofectamin 38. In 2004, Jiao et al studied the transfection of pSecTag2-HCV/NS3, pCI-HCV/NS3, and c3.1-HCV/NS3 into CHO-K1 cells by Lipofectamine. The NS3 expression was detected appropriately 39. In 2008, Lang et al observed that the expression of pConNS3/NS4A was confirmed through transient transfection of a Huh7.0 cell line with Lipofectamine 40.
In 2015, Behzadi et al described the expression of NS3 protein in Huh7 cells using Lipofectamine 41. In the same year, Bolhassani et al showed that TurboFect delivery system increased the efficiency of in vitro transfection of HCV core or coreE1E2 DNA 42. In the current study, the transfection efficiency of both delivery systems was compared (TurboFect and Lipofectamine) for delivering HCV NS3 DNA into HEK-293T cells. In addition, the ability of Hsp20 was evaluated to increase HCV NS3 expression in the cells. Our studies showed that the efficiency of TurboFect transfection reagent was significantly higher than Lipofectamine as a suitable tool for in vitro gene delivery. Moreover, Hsp20 could significantly enhance HCV NS3 DNA delivery and subsequently protein expression in HEK-293T cells using both delivery systems. These results confirm the role of Hsp20 as an antigen carrier.
Conclusion :
Generally, our data showed that the percentage of protein expression using TurboFect was higher than Lipofectamine reagent. In addition, the penetration of Hsp20 into the cells was significantly higher than NS3 using both transfection reagents.
Acknowledgement :
We would like to acknowledge thank Fatemeh Motevalli and Sepideh Shahbazi (Pasteur Institute of Iran) for technical assistance.
Conflict of Interest :
The authors report no conflicts of interest.
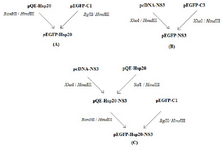
Figure 1. Schematic representation of cloning processes: Generation of pEGFP-Hsp20 (A); pEGFP-NS3 (B); pEGFP-Hsp20-NS3 (C) in bacterial system.
|
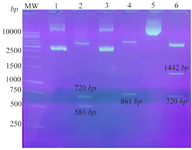
Figure 2. Confirmation of the recombinant plasmids using digestion: Lane 1: pEGFP-Hsp20, Lane 2: pEGFP-Hsp20 digested by NheI/ HindIII (581 bp+720 bp), Lane 3: pEGFP-NS3, Lane 4: pEGFP-NS3 digested by XhoI/HindIII (861 bp), Lane 5: pEGFP-Hsp20-NS3, Lane 6: pEGFP-Hsp20-NS3 digested by NheI/HindIII (1442 bp+720 bp). MW is a molecular weight marker (1 kb, Fermentas).
|
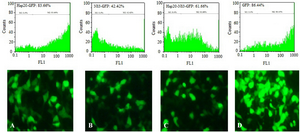
Figure 3. Transfection efficiency: Analysis of Hsp20-GFP (A), NS3-GFP (B), Hsp20-NS3-GFP (C), and GFP (D) protein expression in HEK-293T cells by TurboFect transfection reagent using fluorescent microscopy and flow cytometry. The pEGFP-C1 was used as a positive control (D). The non-transfected cell was considered as a negative control (M1).
|
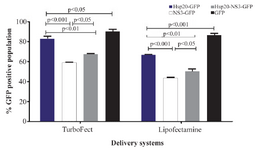
Figure 4. Comparison of TurboFect and Lipofectamine delivery in two independent experiments: The transfection efficiency of Tur-boFect was significantly higher than Lipofectamine.
|
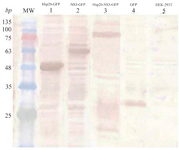
Figure 5. Identification of protein expression in HEK-293T cells 48 hr after transfection using western blot analysis: The expression of Hsp20-GFP (~47 kDa, lane 1), NS3-GFP (~59 kDa, lane 2), and Hsp20-NS3-GFP (~79 kDa, lane 3) was detected by an anti-GFP antibody as compared to the non-transfected cells ( lane 5). The GFP expression (~27 kDa, lane 4) was applied as a positive control. MW is the molecular weight marker.
|
|