Forward Modeling of the Coumarin Antifungals; SPR/SAR Based Perspective
-
Soltani, Saeed
-
Medical Biotechnology Department, Biotechnology Research Center, Pasteur Institute of Iran , Tehran, Iran
-
Dianat, Shima
-
Medical Biotechnology Department, Biotechnology Research Center, Pasteur Institute of Iran , Tehran, Iran
-
 Sardari, Soroush
Ph.D., Medical Biotechnology Department, Biotechnology Research Center, Pasteur Institute of Iran, No. 69, Pasteur Ave, Tehran, Iran, 13164, Tel: +98 21 66405535, Fax: +98 21 66465132, E-mail: ssardari@hotmail.com ; sardari@pasteur.ac.ir
Sardari, Soroush
Ph.D., Medical Biotechnology Department, Biotechnology Research Center, Pasteur Institute of Iran, No. 69, Pasteur Ave, Tehran, Iran, 13164, Tel: +98 21 66405535, Fax: +98 21 66465132, E-mail: ssardari@hotmail.com ; sardari@pasteur.ac.ir
-
Medical Biotechnology Department, Biotechnology Research Center, Pasteur Institute of Iran , Tehran, Iran
Abstract: Although, coumarins are a group of compounds which are naturally found in some plants, they can be synthetically produced as well. Because of their diverse derivatives, origin and properties most of them can be used for medicinal purposes. For example, they can be used against fungal diseases or in studying structure and biological properties of antifungal agents to discover new compounds with the similar activity. A Structure Property/Activity Relationship (SAR) can be utilized in prediction of biological activity of desired molecules.
Introduction :
During the last two decades, human fungal infections have increased among immune compromised individuals (1). Candida albicans (C. albicans) is the major agent of candidosis in humans (2) which is the com-monest invasive fungal infection in patients with malignant haematological disease and in bone marrow transplant recipients (3). One common cause of mortality among hospital-ized patients is nosocomial infection due to opportunistic fungal pathogens (4). The development of azole-based antifungal drugs has revolutionized the treatment of many fungal infections, but therapy may still necessitate application of the highly toxic drug amphotericin B or a combination of drugs. Due to rapid emergence of resistance in fungal pathogens to the conventional drugs, discovery of new potent antifungal com-pounds is necessary. Plant extracts containing coumarin derivatives demonstrate antifungal activity (5) and some synthetic coumarin derivatives are also active against the yeast C. albicans (6). Coumarin is a benzopyrone and a naturally occurring constituent of many plants and essential oils, including tonka beans, sweet clover, woodru, oil of cassia and lavender (7). The presence of phenolic, hydroxy and carboxylic acid groups on the coumarin nucleus has been considered necessary for antimicrobial activity (8). The coumarins are extremely variable in structure and due to the various types of substitutions in the basic structural form their biological activity is influenced (9). As a result, a lot of biological parameters should be evaluated to increase our understanding of the mechanisms by which these coumarins act and a careful structure-property/activity-relationship study of coumarins should be conducted.
The so called "Cheminformatics" was intro-duced to the common use. It is often described as part of the analytical chemistry that by making use of mathematics, probability theory, mathematical statistics, as well as the decision-making theory and computer techniques, has been applied to a diverse range of problems in the field of chemistry (10). By combining to-gether the elements of informatics and chem.-ical analysis, cheminformatics appeared to be particularly useful in the professional work of pharmacists. It is concerned with the search for new chemical compounds as potential drugs, clinical analysis of these compounds, optimiza-tion of drug formulation, evaluation of its quality as well as leading to recognition of complicated processes in which the drug substances are involved in a human organism (11).
Among the multivariate analyses used in the cheminformatics, the principal component analysis (PCA), cluster analysis (CA) and artificial neural networks (ANNs) have been the most widely used methods (12). Their valu-able features are that they can present the correct interpretation of the measured data and obtain the maximum useful information from them (13). A feed-forward Multi-layer Perceptron (MLP) neural network is the most commonly used paradigm in medicinal chem-istry. They usually consist of an input layer, one output layer and one or two hidden or middle layer(s). All units in one layer are connected to all the units in the next layers (14). The signals flow from the first input layer forward through hidden nodes, where a weighed sum of inputs is computed and passed through activation function and the result is finally presented to the output layer. This process is called "feed-forward" (15).
A proper weight setting is not known beforehand and hence, initially, the weights are given a random value. The process of updating the weights to a correct set of values is called "Training or Learning", which is mostly achieved by means of Backpropaga-tion (BP) algorithm (16). The BP is a gen-eralization of the least mean squared algo-rithm that modifies network weight to minimize the mean squared error between the desired and actual outputs of the network. The BP uses supervised learning in which the ne
Materials and Methods :
Data set
The data set was composed of 68 cou-marins and coumarin derivatives selected on the basis of antifungal activity. Antifungal activity of compounds from Table 1 that were screened by the well dilution method has been taken from the literature (20-27).
Authors encountered problems related to reporting of antifungal activity according to the two different forms of minimal inhibitory concentration (MIC) and 50% inhibitory con-centration (IC50) which disabled the analysis of data set with adequate care. To make the dataset uniform, we multiplied the IC50 values by two to obtain a close equivalent of MIC level. Thus, the number generated is approximately equal to MIC for complete inhibition. Preliminary results have shown that coumarins possess considerable anti-fungal activity (5). Therefore, antifungal scre-ening results of isolates of C. albicans were used for the modeling of activity against this microorganism.
Descriptors generation
Eleven attributes have been generated for the description of selected coumarin derive-atives that included eight quantum chemical descriptors; molar refractivity (cm3), molar volume (cm3), parachor (cm3), index of refraction, surface tension (dyne/cm), density (g/cm), polarizability (10-24cm3), molecular mass (Da) and three regular calculated de-scriptors (% carbon, % hydrogen, % oxygen). Calculation of quantum chemical descriptors was preceded by molecular geometry opti-mization based on the PM3 semiempirical approach. Both semiempirical and regular calculations were carried out by ACDLAB 11.02 release 21, May 2008 for in vacuo systems. Besides, quantum chemical descrip-tors, the regular calculated descriptors, % carbon, % hydrogen, and % oxygen) were included in the pool that make better understanding of structure–function activity of coumarin antifungal.
Learning tools
In this study the artificial neural network application of Easy-NNplus 8.0 release 2007, was utilized for SAR model development. Since this technique has been thoroughly described in the reference (28), a detailed description of the method has been omitted. However, a specific implementation of the method for this study is given below.
A standard feed-forward network, with back propagation rule and with one, two or three hidden layer architecture was chosen. The physico-chemical descriptors were used as the inputs, while MIC was the output of the network architecture. In order to avert an over-fitting problem, which is usually pro-duced by more weights due to higher numbers of neurons in input and hidden layers (29), the number of neurons was kept to minimum. However, to produce the optimum architect-ture, powerful enough to model the functions and keep the errors below 0.05%, number of nodes in the hidden layer(s) were varied.
Model validation
Model validation process provides a rea-sonable mean for understanding and approach to molecular design and action mechanism analysis. Applied primary validation methods involved the use of random number gener-ators as a part of the learning process. In order to analyze the influence of inherent random-ness on the prediction stability, ten repetitions of the complete validation process with dif-ferent random seeds were made in all cases (Y-scrambling test). Accuracy has been selec-ted for evaluation of predictive performance of a single validation process, while a cor-relation coefficient (CO) of accuracies obtain-ed across ten repetitions was established as a measure of learning stability. Also cross-validation was applied by leave-n-out method.
Result :
The results of this paper are based on investigation and analysis of collected or calculated data of several coumarin structural descriptors. The artificial neural network system was performed to build a powerful model for prediction of lead and template antifungal coumarins. Table 2 shows results of the various architectures of the neural network system. The numbers of hidden layer nodes were varied according to different node numbers and layers. One of the best architec-tures, considering the correlation behavior and output cycles of calculation was 11-8-4-1. The importance of an input descriptor is determined by the sum of the absolute values of the weights of all the outgoing architecture connections from the input node to the next layer. Some factors, such as surface tension, percent of oxygen, index of refraction, and percentage H have appeared among the most important factors. The least important de-scriptor was determined as the density. A range of predicted activity varied from 125.6796 to 3774.3753. The correlation coef-ficients between the experimental and the predicted MIC value pertaining to all the coumarins was 0.984 (Figure 1).
Compounds 67, 15, and 5 corresponded to the highest error that was generated during the training cycles. Y-Randomization result showed that the classification accuracy for randomized data sets was significantly lower than for the original data sets (data not shown) and hence we concluded that there is no evidence of over-fitting in our models. Cross validation is done by leave-some-out (some= 4) validating method. Validation showed that average of absolute errors was 0.379.
Discussion :
The artificial neural networks (ANNs) have become an important modeling technique in numerous areas of chemistry and pharmacy (30). The mathematical adaptability of ANN commends them as a powerful tool for pattern classification and building predictive models. A particular advantage of ANNs is their inherent ability to incorporate nonlinear de-pendencies between the dependent and in-dependent variables without using an explicit mathematical function.
This study presents an approach to correlate the antifungal activity score data for a data set of drug-like molecules with the structural descriptors. In this study a nonlinear modeling technique of artificial neural network (ANN) with back propagation learning algorithm and sigmoid activation function was used. In this work, a MLP network (29) was developed and used to obtain a nonlinear SAR model. Topo-logically, it consisted of input, hidden, and output layers of neurons or units connected by weights. Each input layer node corresponded to a single independent variable (physico-chemical descriptor) with the exception of the bias node. Similarly, each output layer node corresponded to a different dependent vari-able (property under investigation). In this study, all descriptors were derived solely from molecular structures which did not require experimental data or expensive theoretical calculations (to be obtained).
The ANN model was trained only on the training set since the validation set was used to monitor the external prediction error and thus to avoid overtraining. Among the 11 architectures constructed, the best ANN architecture we found was 11–8–4–1. That is, in the first layer eleven inputs comprised of eleven input descriptors, hidden layer comprised of seven neurons, and the last output layer comprised of one neuron for the property modeled. The statistical criteria obtained for the ANN model are shown in Table 2.
As it can be seen from this table the error for the training set is quite low. In addition, the errors for the validation set are also low showing the good prediction ability. The range of observed and predicted data criterion is very close to each other, that is, the overall prediction is close to experimental. Also, from these result we can conclude that the ANN model satisfactorily predicts the clas-sification nature of the experimental data. Here, we should take into account that a large number of molecular descriptors are usually used in SAR methods. The specific biological action of drugs is frequently described by hydrophobic, electronic, steric and physico-chemical properties. Physicochemical pro-perties characterize the pharmacodynamic properties in the ligand– receptor interaction. They define the ability of the drug to join to the receptor.
The results of this ANN-based study indicate that surface tension is one of the most important factors in coumarin bioactivity. Surface tension of the molecule causes it to creep around the membrane, leading to forma-tion of a layer of loaded molecules at the cell membrane quickly (31). This finding could describe how the LogP is the main sensitivity descriptor of the trained network. Sensitivity analysis is a measure of how the outputs change when the inputs are changed. Result of this paper could help to predict bioactivity of new coumarins.
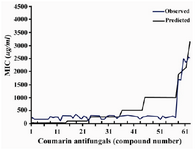
Figure 1. Plot of predicted activity versus the observed one
|

Table 1. Structure and bioactivity of studied coumarins
(*) The observed MICs and structures of coumarin compounds are derived from mentioned references in the table, but predicted MICs have
been calculated by our ANN model.
|
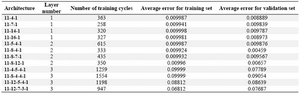
Table 2. Various architecture of neural network and their criteria used in this study
|
|