Generation of a Uracil Auxotroph Strain of the Probiotic Yeast Saccharomyces boulardii as a Host for the Recombinant Protein Production
-
Hamedi, Hassan
-
Department of Food Hygiene, Faculty of Veterinary of Medicine, University of Tehran, Tehran, Iran
-
Misaghi, Ali
-
Department of Food Hygiene, Faculty of Veterinary of Medicine, University of Tehran, Tehran, Iran
-
Zahraei Salehi, Taghi
-
Department of Microbiology, Faculty of Veterinary of Medicine, University of Tehran, Tehran, Iran
-
Khorasanizadeh, Dorsa
-
Fungal Biotechnology Group, Biotechnology Research Center, Pasteur Institute of Iran, Tehran, Iran
-
 Khalaj, Vahid
Fungal Biotechnology Group, Biotechnology Research Center, Pasteur Institute of Iran, Tehran, Iran
Khalaj, Vahid
Fungal Biotechnology Group, Biotechnology Research Center, Pasteur Institute of Iran, Tehran, Iran
-
Fungal Biotechnology Group, Biotechnology Research Center, Pasteur Institute of Iran, Tehran, Iran
Abstract: Background: Saccharomyces boulardii (S. boulardii) is the best known probiotic yeast. The genetic engineering of this probiotic strain requires the availability of appropriate mutants to accept various gene constructs carrying different selection markers. As the auxotrophy selection markers are under focus, we have generated a ura3 auxotroph mutant of S. boulardii for use in further genetic manipulations.
Methods: Classical UV mutagenesis was used for the generation of auxotroph mutants. The mutants were selected in the presence of 5-FOA (5-Fluoroorotic acid), uracil and uridine. Uracil auxotrophy phenotype was confirmed by the ability of mutants to grow in the presence of uracil and the lack of growth in the absence of this compound. To test whether the uracil auxotrophy phenotype is due to the inactivation of URA3 , the mutants were transformed with a plasmid carrying the gene. An in vitro assay was used for the analysis of acid and bile resistance capacity of these mutants
Results: Three mutants were found to be ura3 auxotroph as they were able to grow only in the presence of uracil. When the URA3 gene was added, these mutants were able to grow normally in the absence of uracil. Further in vitro analysis showed that the acid and bile resistance capacity of one of these mutants is intact and similar to the wild type.
Conclusion: A uracil auxotroph mutant of the probiotic yeast, S. boulardii, was generated and characterized. This auxotroph strain may have potential applications in the production and delivery of the recombinant pharmacuetics into the intestinal lumen.
Introduction :
As the knowedge of probiotics expands, more possiblities arise for the engineering of the new probiotic strains. Recombinant probiotics are being considered as efficient biosystems for the delievery of active molecules to the intestinal mucosa 1. S. boulardii is a well known probiotic yeast which is used alone or in combination with probiotic bacteria to support digestive system 2-6. S. boulardii is often marketed in a lyophilized form and is called S. boulardii lyo. The availibility of well-established genetic engineering methods in yeast has facilitated the possible genetic manipulation of this probiotic yeast. In genetic manipulation procedures, the selection of recombinant strains is usually performed by employing a suitable selection marker on a plasmid carrying the gene construct. Antibiotic resistance markers are widely used, but are considered as a major concern in probiotic applications. Hence, it is neseccary to remove the antibiotic resistant gene from the host prior to commercial application 7. In this sense, the auxtrophic markers may be a better substitute as they are indigenous 8. Although these selection markers are commonly used in practice, but they require appropriate host strains which are auxotrophic for the specific nutrients corresponding to the inactivated gene 9. One example of these markers is the URA3 gene that encodes orotidine 5-monophosphate decarboxylase (OMPDCase), an enzyme involved in the de novo synthesis of pyrimidine ribonucleotides 10. The inactiva-tion of URA3 results in uracil auxotrophy and 5-fluoroorotic acid resistance phenotype 11.
In the present study, a uracil auxotroph mu-tant of S. boulardii was generated through UV mutagenesis. The auxotroph mutant was complemented by the URA3 gene. The ura3- mutant strain of S. boulardii can be used in future engineering of this important probiotic yeast.
Materials and Methods :
Strains, media and plasmids: The yeast and bacterial strains used in the present study are listed in table 1. pGEM–T Easy cloning system (Promega) was used for the cloning of PCR products. Plasmid pYES2 (Invitrogen) containing Saccharomyces cerevisiae (S. cerevisiae) URA3 gene was used as a control in transformation experiments.
Yeasts strains were grown and kept in YPD medium (1% yeast extract, 2% polypeptone and 2% dextrose). Yeast Nitrogen Base with ammonium sulphate and without amino acids (YNB medium; Sigma-Aldrich) was prepared at a concentration of 0.67% and was supple-mented with 2% glucose, 10 mM uridine and uracil, and 0.1% 5-FOA (Sigma) to use in screening of auxotrophs.
DNA manipulations: Genomic DNA from both S. cerevisiae Σ1278b and S. boulardii was prepared as de-scribed before 12. All PCRs were performed as 30 cycles of 95oC for 1 min, 58oC for 30 s and 72oC for 1 min. The S. cerevisiae actin fragment (500 bp) was amplified using primers ACT1_F (CCCAATTGAACACGG TATTG) and ACT1_R (GCAGCGGTTTGC ATTTCTTG) as a control in PCR reactions (Table 2).
UV light mutagenesis and isolation of uracil auxotrophs: A single colony from S. boulardii parental strain was grown for 20 hr in YPD broth. Cells were collected and washed with PBS and subjected to UV mutagenesis. 20 ml of cell suspension (1x107 viable yeasts ml-1 in PBS) was gently agitated by a magnetic flea in a glass petri dish (with the lid removed) 15 cm below a UV lamp (Philips, TUV 15W/G15). A dose response experiment was carried out by removing 0.5 ml samples at 10 s intervals over a 100 s period. Irradiated cell suspensions were stored in foil-wrapped tubes at 4°C overnight to avoid photoreactivation. Dilutions of cell suspension from various ex-posure times were made and plated onto YPD agar (3 replicates per dilution). All plates were incubated in the dark at 30°C. Colonies were counted initially after two days and finally after four days of incubation. A kill curve was plotted to estimate the exposure time to UV light to kill 90% of cells. This was then used for the subsequent mutagenesis procedures and the UV irradiated cells were kept at 4°C in a foil-wrapped tube.
To isolate ura3 auxotroph mutants, approximately 107 mutagenized cells were spread onto 5-FOA plates containing uracil and uridine, and then were incubated at 30oC up to one week. The recovered colonies were isolated and plated on YNB medium with or without uracil supplement. Uracil auxotroph mutants were detected by their ability to grow only in the presence of this chemical.
Construction of URA3 cassette: The S. cerevisiae URA3 sequence (URA3/YEL021W, yeast genome database) was used as a template to design the URA3 specific primers. The forward primer, URA3_F (5′-GTT AAT GTG GCT GTG GTT TC-3′), and the reverse primer, URA3_R (5′-GTT ACT TGG TTC TGG CGA GG-3′) (Ta-ble 2), were designed to amplify an approximately 1.2 kb URA3 fragment containing the entire coding sequence with 5` and 3` flanking regions. PCR on genomic DNA of S. boulardii was carried out using these primers. The resulting PCR fragment was cloned into pGEM-Teasy vector. The final vector was called pGEM-ura3 and used in transformation of the auxotroph strains.
Transformation of S. boulardii auxotroph strains: S. boulardii auxotrophic yeasts (ura3-) were transformed with the plasmid pGEM-ura3 us-ing a standard electroporation method 13. As a positive control, a commercial episomal vector, pYES2 (Invitrogen), was used in transformation experiments. Following the trans-formation, cells were plated on YNB agar medium lacking uracil and uridine supplements.
Bile and acid resistance assay: Resistance tests were performed as de-scribed by van der Aa Kühle 14. In brief, all strains were refreshed in MYGP medium (Malt extract 1%, Yeast extract 1%, Peptone 2%, Dextrose 2%) for 24 hr at 30°C. Assays were performed in a 200 µl volume in 96-microwell plates. The wells were inoculated in triplicates with 106 yeast cells and the cells were allowed to grow for 48 hr at 30°C in YNB medium containing acid (pH=2.5) or 0.3% (w/v) Oxgall (Difco). For the auxotroph mutants the medium was supplemented with uracil (10 mM). Viability tests were performed after 4 hr of incubation by plating of 100 µl of cell suspensions onto MYGP agar for 3 days at 30°C.
Results :
UV Survival curve: UV irradiation of cell suspension from S. boulardii was performed and the percentage of survival against time was plotted to esti-mate the UV exposure time required to kill 90% of cells (Figure 1). An exposure time of 23 s was chosen for mutagenesis.
Isolation of URA3 mutants: Mutagenized yeasts were screened for ura3- phenotype on 5-FOA/UU plates. Approximately 350 FOA resistant colonies were isolated and further screened for uracil auxo-trophy (Figure 2). Eight colonies out of 350 were able to grow in YNB-uracil medium but not in YNB. Three mutants, S. boulardii M1, M2 and M3, which had similar growth properties compared to the wild type, were chosen for further studies. These mutants were tested for mutation reversion by plating of 106, 107 and 108 viable cells on YNB agar and counting the number of possible revertants up to 5 days. No revertant was appeared during these incubation periods, indicating that a stable mutation has occurred in the target gene.
Complementation of ura3- mutants of S. boulardii: The URA3 gene from S. boulardii was suc-cessfully amplified as a 1.2 kb fragment using designed specific primers (Figure 3). This was subsequently cloned into pGEM-Teasy vector. The final construct, pGEM-ura3, was confirmed by restriction analysis and se-quencing. The size of URA3 construct was 4.3 kb and the digestion map using SacI/NcoI showed two expected fragments as ~1 kb and 3.3 kb (Figure 4B).
The potential ura3- mutants of S. boulardii (M1, M2 and M3) were transformed with pGEM-ura3 vector. As a positive control, the same mutants were transformed with an epi-somal vector, pYES2, containing URA3 as a selectable marker (Figure 5A). Both trans-formations were efficient and resulted in several hundred transformants from each single reaction (1 µg of each plasmid per reaction).
To confirm that the pYES2 construct is present in these ura3+transformants, plasmid DNA was extracted from one of these transformants and subjected to restriction analysis using EcoRI and ClaI Enzymes. The results confirmed that the isolated plasmid is intact and identical to original plasmid, pYES2 (Figure 5B).
Acid and bile resistance in auxotroph mutants: The ability of auxotroph mutants to resist pH=2.5 and 0.3% oxgall was assessed. Table 3 shows the growth phenotype of three different mutants compared to the wild types. All tested strains showed different resistance against acid and bile. Among the mutants, only S. boulardii M2 showed a resistance pattern similar to the S. boulardii wild type.
Discussion :
The design, creation and genetic manipulation of probiotic strains exclusively as vaccine and drug delivery vehicles are promising and rapidly growing area of research 1,15.
The yeast S. boulardii can be considered as a candidate probiotic for future engineering. To facilitate the genetic manipulation of this yeast, we used the classical UV mutagenesis to produce uacil auxotroph mutants of S. boulardii as a host for recombinant protein production. The UV dose-response curve demonstrated a 90% killing rate after 23 s of UV irradiation. This result is in agreement with the time range reported by Hashimoto et al (20-40S) 7.
Ura3- mutants were selected on 5-FOA plates. 5-FOA is toxic to yeast cells that can synthesize the ura3 gene product, and therefor makes them unable to grow on 5-FOA-containing media 11. In addition to act as a positive selection marker, the URA3 gene can also be used for the negative selection (counter selection). In this regard, the presence of URA3 confers sensitivity to FOA, while ura3- negative cells are FOA resistant. This concept has been used in designing the ura-blaster gene constructs as a tool in multiple gene disruption experiments in S. cerevisiae 16. Hence, the generation of uracil auxotrophs of S. boulardii provides an opportunity for gene deletion studies in this organism.
To complement the ura3- phenotype, the URA3 gene was amplified from the S. boulardii genome and cloned into pGEM-Teasy vector. The restriction analysis of the URA3 fragment from S. boulardii showed a pattern identical to its homologue in S. cerevisiae. This pattern was expected as the analysis of sequence data from different strains of S. boulardii had confirmed a high similarity between S. boulardii and S. cerevisiae in DNA level 17.
Bile and acid resistance are the most important prerequisites for probiotics to stay alive in the digestive tract of their hosts. Among the three isolated ura3- mutants, only one (S. boulardi M2) showed acceptable resistance to acid and bile. Similarly, Sharaf et al. have used EMS mutagenesis and interspecific protoplast fusion to isolate improved probiotic yeasts. They isolated an adenine auxotroph mutant of S. boulardii with high tolerance to bile salt 18. Abosereh et al have also isolated highly resistant S. boulardii strains through protoplast fusion 19. The acid and bile resitance capability of mutants provide these strains with an advantage in vivo. Further in vivo studies are underway to evaluate other probiotic features of the mutant.
Conclusion :
A uracil auxotroph mutant of the probiotic yeast, S. boulardii, was generated in this study. The mutant was complemented by URA3 carrying constructs, confirming the inactivation of this gene in the mutant. Bile and acid resistance of the mutant was the same as wild type strain. This mutant can be used as a probiotic host for the in vivo production and delivery of various recombinant products to the GI tract.
Acknowledgement :
This study was financially supported by a grant awarded to A. Misaghi. All scientific experiments were carried out in VK lab at Pasteur Institute of Iran.
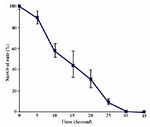
Figure 1. Survival rate of S.boulardii following UV irradiation. Cell counts were performed in triplicates
|
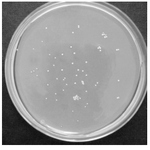
Figure 2. FOA resistant colonies on YNB plates. Approximately 107 mutagenized cells (10% survival) were plated on YNB-FOA -UU plates. The resistant colonies appeared after 5-7 days
|
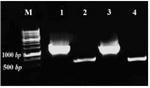
Figure 3. Amplification of ura3 and actin fragments using genomic DNA of S. cerevisiae and S. boulardii. M: Size marker, 1: S. cerevisiae ura3 fragment (1.2 kb), 2: S. cerevisiae actin fragment (0.5 kb), 3: S. boulardii ura3 and 4: S. boulardii actin fragments
|
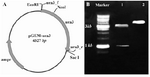
Figure 4. A) Schematic representation of pGEM-ura3 construct. The position of designed primers and restriction sites is shown. B) Restriction analysis of pGEM-ura3 vector. Lane 1: Fragments generated by NcoI/SacI digestion of the construct (~1 kb and ~3.3 kb). Lane 2: NcoI linearized plasmid
|
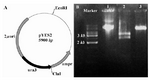
Figure 5. A) Schematic representation of pYES2 vector. pYES2 contains the auxotrophic marker URA3, 2 µ origin and the ampicilin resistance marker. B) Restriction analysis of pYES2. Lane 1: undigested plasmid. Lane2: EcoRI/ClaI digested vector showed two expected bands of ~2.5 and ~3.2 kb. Lane 3: ClaI linearized plasmid (5.9 kb)
|
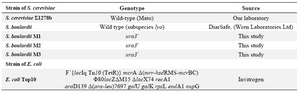
Table 1. Strains used in this study
|
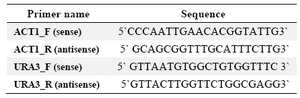
Table 2. Primers used in this study
|
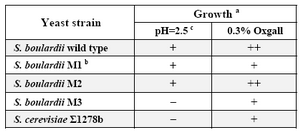
Table 3. Growth ability of wild type strains and ura3 mutants in the presence of acid and bile
A) -no growth; + growth delay>4 hr; ++no delay in growth.
B) M1, M2 and M3:the ura3 mutant of S. boulardii
C) Survival after 4-hr incubation at pH= 2.5.
|
|