Fetal Sex Determination using Non-Invasive Method of Cell-free Fetal DNA in Maternal Plasma of Pregnant Women During 6th– 10th Weeks of Gestation
-
Zargari, Maryam
-
Biology Department, Science and Research Branch, Islamic Azad University (IAU), Tehran, Iran
-
Sadeghi, Mohammad Reza
-
Reproductive Biotechnology Research Centre, Avicenna Research Institute, ACECR, Tehran, Iran
-
Kamali, Koorosh
-
Reproductive Biotechnology Research Centre, Avicenna Research Institute, ACECR, Tehran, Iran
-
Saliminejad, Kyomars
-
Reproductive Biotechnology Research Centre, Avicenna Research Institute, ACECR, Tehran, Iran
-
Esmaeilzadeh, Ali
-
Reproductive Biotechnology Research Centre, Avicenna Research Institute, ACECR, Tehran, Iran
-
 Khorram Khorshid, Hamid Reza
Genetic Research Centre, University of Social Welfare and Rehabilitation Sciences, Tehran, Iran, Avicenna Research Institute (ACECR), Shahid Beheshti University, Evin, Tehran, Iran, Tel: +98 21 22180138 Fax: +98 21 22432020 E-mail: hrkk1@uswr.ac.ir
Khorram Khorshid, Hamid Reza
Genetic Research Centre, University of Social Welfare and Rehabilitation Sciences, Tehran, Iran, Avicenna Research Institute (ACECR), Shahid Beheshti University, Evin, Tehran, Iran, Tel: +98 21 22180138 Fax: +98 21 22432020 E-mail: hrkk1@uswr.ac.ir
-
Reproductive Biotechnology Research Centre, Avicenna Research Institute, ACECR, Tehran, Iran
-
Genetic Research Centre, University of Social Welfare and Rehabilitation Sciences, Tehran, Iran
Abstract: In previous years, identification of fetal cells in maternal blood circulation has caused a new revolution in non-invasive method of prenatal diagnosis. Low number of fetal cells in maternal blood and long-term survival after pregnancy limited the use of fetal cells in diagnostic and clinical applications. With the discovery of cell-free fetal DNA (cffDNA) in plasma of pregnant women, access to genetic material of the fetus had become possible to determine early gender of a fetus in pregnancies at the risk of X-linked genetic conditions instead of applying invasive methods. Therefore in this study, the probability of detecting sequences on the Y chromosome in pregnant women has been evaluated to identify the gender of fetuses. Peripheral blood samples were obtained from 80 pregnant women at 6th to 10th weeks of gestation and then the fetal DNA was extracted from the plasma. Nested PCR was applied to detect the sequences of single copy SRY gene and multi copy DYS14 & DAZ genes on the Y chromosome of the male fetuses. At the end, all the obtained results were compared with the actual gender of the newborns. In 40 out of 42 born baby boys, the relevant gene sequences were identified and 95.2% sensitivity was obtained.
Conclusion: Non-invasive early determination of fetal gender using cffDNA could be employed as a pre-test in the shortest possible time and with a high reliability to avoid applying invasive methods in cases where a fetus is at the risk of genetic diseases.
Introduction :
Traditionally, early fetal gender determination has been performed using invasive techniques, such as chorionic villus sampling or amniocentesis. These procedures, however, still carry a risk of miscarriage around 1-2% and cannot be performed until 11 weeks of gestation (1). Also, reliable determination of fetal sex by means of ultrasonography cannot be done in the first trimester because of uncompleted development of the external genitalia (2).
Therefore, more efforts have been spent in developing prenatal diagnostic procedures that do not constitute a risk for the fetus, based on the analysis of fetal genetic material obtained from the fetal cells circulating in maternal blood (3,4). Substantial advances have been made in the enrichment and isolation of fetal cells for analysis, but most techniques are time-consuming or require expensive equipment. In addition, these cells are very rare in maternal plasma (1 fetal cell per 106 maternal cells) and they are unlikely to persist after delivery, including subsequent pregnancies (5,6). Further studies on tumour derived DNAs in the plasma of cancer patients open up the possibility that fetal DNA which originated from apoptotic trophoblasts of the placenta, may also be found in maternal plasma (7,8). Finally with the discovery of cell-free fetal DNA (cffDNA) fragments in the plasma of pregnant women carrying male fetus in 1997, reliable and accurate diagnosis became reality (9).
More studies revealed that the concentration of fetal DNA in maternal plasma was found to be much higher than that present in the cellular fraction (25.4 GEq/ml in early stage of pregnancy and 292.2 GEq/ml in late stage of pregnancy) (10) and the post-partum clearance of cffDNA from the maternal circulation was rapid with a mean half-life of 16.3 min (11). Also, fetal DNA molecules are generally shorter than maternal DNA molecules (between 193 bp to 313 bp). Therefore, it can be distinguished from the maternal DNA by size separation (12). These findings have provided the opportunity to perform reliable genetic testing on cffDNA extracted from the maternal plasma at an early stage in pregnancy without interference from previous pregnancies for non-invasive prenatal diagnosis of paternally inherited disorders as well as fetal gender determination.
Using cffDNA in maternal plasma for fetal gender determination is mainly limited to those sequences which are absent from the maternal genome such as the SRY, DYS14 and DAZ that are located on the Y chromosome. Therefore, the only way to identify these sequences is through male-bearing pregnancies (13).
In this study, early determination of fetal gender using cffDNA can be considered as a non-invasive pre-test to determine whether invasive prenatal diagnosis should be performed on a fetus having a risk of X-linked disorders or not. Thus, invasive procedures can be avoided when the fetus is known to be female at an early gestational age, while prenatal diagnosis might be performed only for male fetuses. To achieve this goal, the following study using cffDNA in maternal plasma was performed on pregnant women during their 6th -10th weeks of pregnancy to obtain the required sensitivity, specificity and accuracy for a non-invasive prenatal test.
Materials and Methods :
Sample collection
Peripheral blood samples were obtained from 80 pregnant women at their 6th to 10th weeks of gestation who were referred to Avicenna Infertility Clinic in Tehran, Iran during 2009-2010. Also in this study, five non-pregnant women and five men were considered as a negative and positive control. Before blood sampling, signed consent forms were obtained from all participants and the protocol of the study was approved by the ethics committee of Avicenna research institute. For each case, 5 ml peripheral blood was collected in a tube containing 200 µl of 0.5 M EDTA and immediately stored at 4C. Within 24 hr after collection, blood samples were centrifuged at 3000 g for 10 min and the upper plasma layer was carefully removed without disturbing the buffy coat, transferred into a new Eppendorf tube for storage at
-20C until further processing.
DNA Extraction
Genomic DNA was extracted from 200 µl of the plasma samples using the QIAamp DNA Blood Mini kit (Qiagen, USA) as recommended by the manufacturer according to the manufacturer’s “Blood and Body Fluid” protocol. The extracted DNA was eluted in 50 μl of the elution buffer.
Primers
Due to the low concentration of cffDNA in maternal plasma, multicopy DAZ and DYS14 genes were used for detection of sequences on the Y chromosome in male-bearing pregnancies. Also, sequence of single copy SRY gene was used as an internal control of gender determination. In order to assess the presence of sufficient cell-free DNA in an extraction, analysis of the ACTB sequences was performed. All the pairs of primers were designed by using the Primer3 and Gene Runner software (Table 1).
Nested PCR
The fetal sex was identified by nested PCR technique to amplify the DYS14 and DAZ sequences, whereas the semi-nested PCR was used for amplification of SRY sequences. In the first round of PCR, the amplification reactions were set in a total volume of 25 μl containing: 2 μL of 10 ng genomic DNA, 2.5 μL of 10X PCR buffer (Roch, Germany), 1 μl of 10 mM dNTPs, 10 pmol of each primers, 1 unit of Taq DNA polymerase (Roch, Germany), 2.5 μl of Mgcl2 (for SRY and DAZ) and a 3 μl of Mgcl2 for DYS14. Amplification was performed using a programmable thermal cycler gradient PCR system (Eppendorf, Germany) with an initial denaturation of 94°C for 5 min followed by 40 cycles of denaturation at 94°C for 30 sec, annealing at 62°C for 30 sec (for SRY, DYS14) and 64°C (for DAZ), extension at 72°C for 30 sec and the final extension at 72°C for 10 min.
One μl of the PCR products were used as template for the second round of PCR. The reaction system and thermo-cycling condition were the same as those of the first round of PCR but they were carried out in 35 cycles. At the end of each round of PCR, amplification products were visualized by staining with ethidium bromide after electrophoresis on 2% agarose gel. To recognize contamination, a non template control, containing DNA free water was also added in each reaction.
Anti-contamination measures
As an anti-contamination measure, an aerosol-resistant pipette tips were used for all liquids and separate areas were considered for all the steps of the analysis. In order to eliminate contact with exogenous male DNA, only female operators were selected in all procedures using a laminar flow hood.
Statistical analysis
At the end, all the obtained results were compared with the actual gender of the newborns to calculate the sensitivity, specificity and accuracy of the applied method. Kappa coefficient of agreement was calculated to evaluate the precision and correctness of the method and for evaluation of the clinical application, parameters such as Positive Predictive Value (PPV) and Negative Predictive Value (NPV) were also analysed. Statistical analysis was performed using the SPSS statistical package (V.11.5).
Results :
Statistical analysis of the data showed that from the total of 80 samples obtained from pregnant women with age range of 18 to 46 years (Mean age=30.36) and pregnancy week range from 6 to 10 weeks (Mean week=7.74), abortion was observed in 5 cases. Although the gender determination tests were conducted on these 5 samples, they were excluded from statistical analysis due to lack of knowledge of the actual fetal gender to compare with test results. Also, among the 80 samples, 16 cases with multiple gestations (9 identical twins, 6 non-identical twins and 1 triplet) were observed which were included in our statistical analysis.
Nested PCR results
In all the obtained samples, ACTB sequences were amplified (PCR product length=149 bp) indicating the presence of sufficient DNA in the extracted samples (Figure 1A). Regarding the SRY, DYS14 and DAZ sequences, no positive bands of PCR were observed in samples during the first round of PCR. However, on the electrophoresis results of the second round of PCR (Figure 1B, C, and D), positive bands (PCR product length=143 bp, 122 bp and 156 bp, respectively) were observed for each SRY, DYS14 and DAZ sequences in 40 samples similar to positive control DNA sample which was recorded as positive, and as a result the male gender. On the other hand, in the absence of positive bands of PCR for 2 or 3 of the mentioned sequences in fetal samples similar to negative control DNA sample, they were considered as negative and would represent the female gender. It should be mentioned that in all the 40 samples classified as male fetuses all the 3 marker targets were positive.
Comparison of the obtained results with the actual birth outcome indicates that from 50 baby girls born, 49 fetal genders were correctly diagnosed and only in one case, positive result was obtained. Also from 42 baby boys born, 40 genders were correctly diagnosed by using the SRY, DYS14 and DAZ sequences (Table 2).
The sensitivity and specificity of the method used in fetal gender determination (with 95% confidence intervals) were respectively 95.2% (95% CI= 0.842 to 0.987) and 98% (95% CI= 0.895 to 0.996). Kappa coefficient of agreement in relevant test is 0.934 and the p-values=0.001 were considered statistically significant in each analysis using SRY, DYS14 and DAZ sequences. The PPV and NPV of the method used respectively were 97.6% (95% CI= 0.895-0.996) and 96.1% (95% CI=0.868-0.989).
Discussion :
In recent years, the existence of circulating fetal cell-free DNA in maternal blood which was discovered by Lo et al in 1997, created an outstanding revolution in non-invasive prenatal diagnosis. Fetal nucleic acids can be obtained from a simple blood draw of the mother that is risk-free and highly cost-effective compared to conventional invasive methods. Despite the low concentration of cffDNA (3.4% to 6.2% of total maternal DNA) and with the advent of molecular techniques such as Polymerase Chain Reaction (PCR), nucleic acid based testing has become a valuable source for non-invasive prenatal diagnosis since nucleic acids can be amplified (10). Therefore, in this study by using nested PCR technique, 95.2% sensitivity was achieved in identification of male fetuses using SRY, DYS14 and DAZ sequences. This is very close to the sensitivity obtained in previous studies. And in most cases, even significantly higher sensitivity was obtained.
Previous investigations have shown sensitivity of 94% (Smid et al, 1999), 96% (Al-Yatama et al, 2001), 94% (Zolotukhina et al, 2005) and 88.2% (Hong et al, 2006) (14-17). However in these findings, longer time range of pregnancy were considered, which led to high probability of identifying cffDNA because of the gradual increase of its concentration with increasing gestational age (18). In comparison, the results obtained in this study in the 6th to 10th weeks of pregnancy led to significantly better results. Overall, the differences observed between the results of the previous studies and this study can be explained through the use of different methods of fetal DNA extraction, low concentration of cffDNA at an early gestational age, number of population, time range of the pregnancy and existence of potential contamination.
In this study, false positive result was observed only in one case that gender of female fetus, which was at the age of six weeks, was diagnosed as a male. Citing the theory of vanishing twins within the first 7 weeks of gestation in 0.3-0.7% of pregnancies (19), it can be concluded that during the time of sampling in the 6th week of gestation, there was a male twin that disappeared in the subsequent weeks of pregnancy and only the baby girl was born. On the other hand, false negative results were obtained in two cases of pregnancies with non-identical twins which can be the result of extraction failure of the male twin DNA compared to the female twin due to its low concentration of cffDNA. Therefore if we do not consider multiple gestations in our study, we have reached a significant 100% sensitivity.
In comparison between total numbers of fetuses that were correctly diagnosed and total numbers of infant born, 96.7% accuracy was achieved in fetal gender determination. Also according to the Kappa coefficient of agreement, which is in “almost perfect” agreement range between 0.81 and 0.99 (20), it can be concluded that the results are remarkably consistent with the actual gender of the babies.
To check whether the method used in this study is suitable for clinical application or not, the parameters of PPV and NPV were calculated. The PPV and NPV indicate that if the test results are positive and the fetus is diagnosed as a boy, there is 97.6% probability to be a boy and 96.1% probability to be a girl if the test results are negative. Therefore, this method can be used as a clinical method in determining the fetal gender due to its high probability of correct prediction, prior to applying invasive methods.
Conclusion :
We hope that non-invasive fetal gender determination using cffDNAs in maternal plasma would allow us to obtain an early knowledge of the fetal sex and adding to timely clinical management. This could reduce the need for invasive procedures in pregnant women carrying an X-linked disorder up to 50%. Also, hopefully in the near future, this method can be applied as a diagnostic tool for diseases of pregnancy such as preeclampsia or preterm labor, or for fetal anomalies such as aneuploidies.
Acknowledgement :
We would like to thank all of the individuals who kindly accepted to participate in this study. Also, we are deeply indebted to the personnel of Avicenna Infertility Clinic for their assistance in sample collection and to Ms. H. Edalatkhah for their helpful advices. Financially, this study was supported by Avicenna Research Institute.
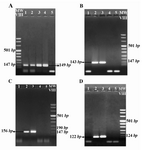
Figure 1. The agarose gel electrophoresis result of ACTB (A), SRY (B), DYS14 (C) and DAZ (D) amplification by nested PCR analysis in fetal DNA samples extracted from the maternal plasma. Lanes 1 & 2: Fetal DNA, Lane 3: Male DNA as a positive control, Lane 4: non pregnant women DNA as a negative control and Lane 5: PCR reaction negative Control (Water). A) 149 bp Positive bands in lane 1 and 2 indicate the presence of sufficient DNA in the extracted samples. Observation of positive bands 143 bp B) 122 bp C) and 156 bp D) in lane 2 shows the amplification of the relevant genes and indicate that the gender of the fetus is male. The absence of positive band in lane 1 indicates the female gender of the fetus
|

Table 1. Primer sequences used in PCR
|

Table 2. Comparison of test results by nested PCR with the actual birth outcome in 80 samples during 6th to 10th weeks of gestation
a False positive result
b False negative result
|
|